Gadd45, a p53-responsive stress protein, modifies DNA accessibility on damaged chromatin
- PMID: 10022855
- PMCID: PMC83961
- DOI: 10.1128/MCB.19.3.1673
Gadd45, a p53-responsive stress protein, modifies DNA accessibility on damaged chromatin
Abstract
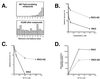
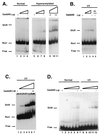
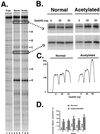
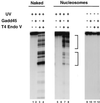
This report demonstrates that Gadd45, a p53-responsive stress protein, can facilitate topoisomerase relaxing and cleavage activity in the presence of core histones. A correlation between reduced expression of Gadd45 and increased resistance to topoisomerase I and topoisomerase II inhibitors in a variety of human cell lines was also found. Gadd45 could potentially mediate this effect by destabilizing histone-DNA interactions since it was found to interact directly with the four core histones. To evaluate this possibility, we investigated the effect of Gadd45 on preassembled mononucleosomes. Our data indicate that Gadd45 directly associates with mononucleosomes that have been altered by histone acetylation or UV radiation. This interaction resulted in increased DNase I accessibility on hyperacetylated mononucleosomes and substantial reduction of T4 endonuclease V accessibility to cyclobutane pyrimidine dimers on UV-irradiated mononucleosomes but not on naked DNA. Both histone acetylation and UV radiation are thought to destabilize the nucleosomal structure. Hence, these results imply that Gadd45 can recognize an altered chromatin state and modulate DNA accessibility to cellular proteins.
Figures


References
-
- Antinore, M. J. Unpublished data.
-
- Bae I, Smith M L, Sheikh M S, Zhan Q, Scudiero D A, Friend S H, O’Connor P M, Fornace A J., Jr An abnormality in the p53 pathway following γ-irradiation in many wild-type p53 human melanoma lines. Cancer Res. 1996;56:840–847. - PubMed
-
- Bhatia K, Pommier Y, Giri C, Fornace A J, Jr, Imaizumi M, Breitman T R, Cherney B W, Smulson M E. Expression of the poly(ADP-ribose) polymerase gene following natural and induced DNA strand breakage and effect of hyperexpression on DNA repair. Carcinogenesis. 1990;11:123–128. - PubMed
-
- Bianchi M E, Beltrame M, Paonessa G. Specific recognition of cruciform DNA by nuclear-protein HMG1. Science. 1989;243:1056–1059. - PubMed
-
- Boffa L C, Vidali G, Mann R S, Allfrey V G. Suppression of histone deacetylation in vivo and in vitro by sodium butyrate. J Biol Chem. 1978;253:3364–3366. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
