The Jun kinase 2 isoform is preferentially required for epidermal growth factor-induced transformation of human A549 lung carcinoma cells
- PMID: 10022881
- PMCID: PMC83987
- DOI: 10.1128/MCB.19.3.1938
The Jun kinase 2 isoform is preferentially required for epidermal growth factor-induced transformation of human A549 lung carcinoma cells
Abstract
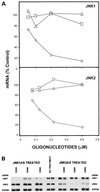
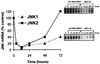
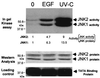
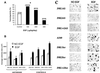
We have previously found that epidermal growth factor (EGF) mediates growth through the Jun N-terminal kinase/stress-activated kinase (JNK/SAPK) pathway in A549 human lung carcinoma cells. As observed here, EGF treatment also greatly enhances the tumorigenicity of A549 cells, suggesting an important role for JNK in cancer cell growth (F. Bost, R. McKay, N. Dean, and D. Mercola, J. Biol. Chem. 272:33422-33429, 1997). Several isoforms families of JNK, JNK1, JNK2, and JNK3, have been isolated; they arise from alternative splicing of three different genes and have distinct substrate binding properties. Here we have used specific phosphorothioate oligonucleotides targeted against the two major isoforms, JNK1 and JNK2, to discriminate their roles in EGF-induced transformation. Multiple antisense sequences have been screened, and two high-affinity and specific candidates have been identified. Antisense JNK1 eliminated steady-state mRNA and JNK1 protein expression with a 50% effective concentration (EC50) of <0.1 microM but did not alter JNK2 mRNA or protein levels. Conversely, antisense JNK2 specifically eliminated JNK2 steady-state mRNA and protein expression with an EC50 of 0.1 microM. Antisense JNK1 and antisense JNK2 inhibited by 40 and 70%, respectively, EGF-induced total JNK activity, whereas sense and scrambled-sequence control oligonucleotides had no effect. The elimination of mRNA, protein, and JNK activities lasted 48 and 72 h following a single Lipofectin treatment with antisense JNK1 and JNK2, respectively, indicating sufficient duration for examining the impact of specific elimination on the phenotype. Direct proliferation assays demonstrated that antisense JNK2 inhibited EGF-induced doubling of growth as well as the combination of active antisense oligonucleotides did. EGF treatment also induced colony formation in soft agar. This effect was completely inhibited by antisense JNK2 and combined-antisense treatment but not altered by antisense JNK1 alone. These results show that EGF doubles the proliferation (growth in soft agar as well as tumorigenicity in athymic mice) of A549 lung carcinoma cells and that the JNK2 isoform but not JNK1 is utilized for mediating the effects of EGF. This study represents the first demonstration of a cellular phenotype regulated by a JNK isoform family, JNK2.
Figures




References
-
- Adamson E D, Wiley L M. The EGFR gene family in embryonic cell activities. Curr Top Dev Biol. 1997;35:71–120. - PubMed
-
- Adler V, Polotskaya A, Kim J, Dolan L, Davis R, Pincus M, Ronai Z. Dose rate and mode of exposure are key factors in JNK activation by UV irradiation. Carcinogenesis. 1996;9:2073–2076. - PubMed
-
- Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochem Biophys Acta. 1991;1072:129–157. - PubMed
-
- Antonyak M A, Moscatello D K, Wong A J. Constitutive activation of c-Jun N-terminal kinase by a mutant epidermal growth factor receptor. J Biol Chem. 1998;273:2817–2822. - PubMed
-
- Baldassarre G, Bianco C, Tortora G, Ruggiero A, Moasser M, Dmitrovsky E, Bianco A R, Ciardiello F. Transfection with a cripto antisense plasmid suppresses endogenous cripto expression and inhibits transformation in a human embryonal carcinoma cell line. Int J Cancer. 1996;66:538–543. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
