Different regulation of the p53 core domain activities 3'-to-5' exonuclease and sequence-specific DNA binding
- PMID: 10022902
- PMCID: PMC84008
- DOI: 10.1128/MCB.19.3.2155
Different regulation of the p53 core domain activities 3'-to-5' exonuclease and sequence-specific DNA binding
Abstract
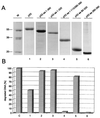
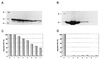
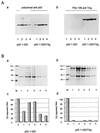
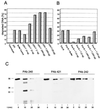
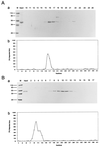
In this study we further characterized the 3'-5' exonuclease activity intrinsic to wild-type p53. We showed that this activity, like sequence-specific DNA binding, is mediated by the p53 core domain. Truncation of the C-terminal 30 amino acids of the p53 molecule enhanced the p53 exonuclease activity by at least 10-fold, indicating that this activity, like sequence-specific DNA binding, is negatively regulated by the C-terminal basic regulatory domain of p53. However, treatments which activated sequence-specific DNA binding of p53, like binding of the monoclonal antibody PAb421, which recognizes a C-terminal epitope on p53, or a higher phosphorylation status, strongly inhibited the p53 exonuclease activity. This suggests that at least on full-length p53, sequence-specific DNA binding and exonuclease activities are subject to different and seemingly opposing regulatory mechanisms. Following up the recent discovery in our laboratory that p53 recognizes and binds with high affinity to three-stranded DNA substrates mimicking early recombination intermediates (C. Dudenhoeffer, G. Rohaly, K. Will, W. Deppert, and L. Wiesmueller, Mol. Cell. Biol. 18:5332-5342), we asked whether such substrates might be degraded by the p53 exonuclease. Addition of Mg2+ ions to the binding assay indeed started the p53 exonuclease and promoted rapid degradation of the bound, but not of the unbound, substrate, indicating that specifically recognized targets can be subjected to exonucleolytic degradation by p53 under defined conditions.
Figures


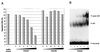



References
-
- Bargonetti J, Manfredi J J, Chen X, Marshak D R, Prives C. A proteolytic fragment from the central region of p53 has marked sequence-specific DNA-binding activity when generated from wild-type but not from oncogenic mutant p53 protein. Genes Dev. 1993;7:2565–2574. - PubMed
-
- Bargonetti J, Reynisdottir I, Friedman P N, Prives C. Site-specific binding of wild-type p53 to cellular DNA is inhibited by SV40 T antigen and mutant p53. Genes Dev. 1992;6:1886–1898. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
