The highly conserved beta-hairpin of the paired DNA-binding domain is required for assembly of Pax-Ets ternary complexes
- PMID: 10022910
- PMCID: PMC84016
- DOI: 10.1128/MCB.19.3.2231
The highly conserved beta-hairpin of the paired DNA-binding domain is required for assembly of Pax-Ets ternary complexes
Abstract
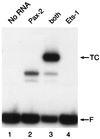
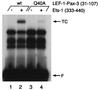
Pax family transcription factors bind DNA through the paired domain. This domain, which is comprised of two helix-turn-helix motifs and a beta-hairpin structure, is a target of mutations in congenital disorders of mice and humans. Previously, we showed that Pax-5 (B-cell-specific activator protein) recruits proteins of the Ets proto-oncogene family to bind a composite DNA site that is essential for efficient transcription of the early-B-cell-specific mb-1 promoter. Here, evidence is provided for specific interactions between Ets-1 and the amino-terminal subdomains of Pax proteins. By tethering deletion fragments of Pax-5 to a heterologous DNA-binding domain, we show that 73 amino acids (amino acids 12 to 84) of its amino-terminal subdomain can recruit the ETS domain of Ets-1 to bind the composite site. Furthermore, an amino acid (Gln22) within the highly conserved beta-hairpin motif of Pax-5 is essential for efficient recruitment of Ets-1. The ability to recruit Ets proteins to bind DNA is a shared property of Pax proteins, as demonstrated by cooperative DNA binding of Ets-1 with sequences derived from the paired domains of Pax-2 and Pax-3. The strict conservation of sequences required for recruitment of Ets proteins suggests that Pax-Ets interactions are important for regulating transcription in diverse tissues during cellular differentiation.
Figures





Similar articles
-
Highly conserved amino acids in Pax and Ets proteins are required for DNA binding and ternary complex assembly.Nucleic Acids Res. 2001 Oct 15;29(20):4154-65. doi: 10.1093/nar/29.20.4154. Nucleic Acids Res. 2001. PMID: 11600704 Free PMC article.
-
Requirements for selective recruitment of Ets proteins and activation of mb-1/Ig-alpha gene transcription by Pax-5 (BSAP).Nucleic Acids Res. 2003 Oct 1;31(19):5483-9. doi: 10.1093/nar/gkg785. Nucleic Acids Res. 2003. PMID: 14500810 Free PMC article.
-
Pax-5 (BSAP) recruits Ets proto-oncogene family proteins to form functional ternary complexes on a B-cell-specific promoter.Genes Dev. 1996 Sep 1;10(17):2198-211. doi: 10.1101/gad.10.17.2198. Genes Dev. 1996. PMID: 8804314
-
Ets-1 flips for new partner Pax-5.Structure. 2002 Jan;10(1):11-4. doi: 10.1016/s0969-2126(01)00701-8. Structure. 2002. PMID: 11796106 Review.
-
Regulation of Ets function by protein - protein interactions.Oncogene. 2000 Dec 18;19(55):6514-23. doi: 10.1038/sj.onc.1204035. Oncogene. 2000. PMID: 11175367 Review.
Cited by
-
PAX3 and ETS1 synergistically activate MET expression in melanoma cells.Oncogene. 2015 Sep 17;34(38):4964-74. doi: 10.1038/onc.2014.420. Epub 2014 Dec 22. Oncogene. 2015. PMID: 25531327 Free PMC article.
-
Characterization of seven genes affecting Caenorhabditis elegans hindgut development.Genetics. 1999 Oct;153(2):731-42. doi: 10.1093/genetics/153.2.731. Genetics. 1999. PMID: 10511553 Free PMC article.
-
Establishment of the lymphoid ETS-code reveals deregulated ETS genes in Hodgkin lymphoma.PLoS One. 2023 Jul 10;18(7):e0288031. doi: 10.1371/journal.pone.0288031. eCollection 2023. PLoS One. 2023. PMID: 37428779 Free PMC article.
-
Molecular mechanisms of ETS transcription factor-mediated tumorigenesis.Crit Rev Biochem Mol Biol. 2013 Nov-Dec;48(6):522-43. doi: 10.3109/10409238.2013.838202. Epub 2013 Sep 25. Crit Rev Biochem Mol Biol. 2013. PMID: 24066765 Free PMC article. Review.
-
Comparative analysis of the mammalian WNT4 promoter.BMC Genomics. 2009 Sep 6;10:416. doi: 10.1186/1471-2164-10-416. BMC Genomics. 2009. PMID: 19732466 Free PMC article.
References
-
- Adams B, Dorfler P, Aguzzi A, Kozmik Z, Urbanek P, Maurer-Fogy I, Busslinger M. Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS, and adult testis. Genes Dev. 1992;6:1589–607. - PubMed
-
- Asher J H, Jr, Sommer A, Morell R, Friedman T B. Missense mutation in the paired domain of PAX3 causes craniofacial-deafness-hand syndrome. Human Mutat. 1996;7:30–35. - PubMed
-
- Balczarek K A, Lai Z-C, Kumar S. Evolution and functional diversification of the paired box (Pax) DNA-binding domains. Mol Biol Evol. 1997;14:829–842. - PubMed
-
- Baldwin C T, Hoth C F, Amos J A, daSilva E O, Milunsky A. An exonic mutation in the HuP2 paired domain gene causes Waardenburg’s syndrome. Nature. 1992;355:637–638. - PubMed
-
- Balling R, Deutsch U, Gruss P. undulated, a mutation affecting the development of the mouse skeleton, has a point mutation in the paired box of Pax-1. Cell. 1988;55:531–535. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
