Identification of a novel family of targets of PYK2 related to Drosophila retinal degeneration B (rdgB) protein
- PMID: 10022914
- PMCID: PMC84020
- DOI: 10.1128/MCB.19.3.2278
Identification of a novel family of targets of PYK2 related to Drosophila retinal degeneration B (rdgB) protein
Abstract
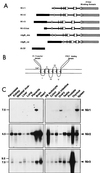
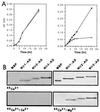
The protein tyrosine kinase PYK2 has been implicated in signaling pathways activated by G-protein-coupled receptors, intracellular calcium, and stress signals. Here we describe the molecular cloning and characterization of a novel family of PYK2-binding proteins designated Nirs (PYK2 N-terminal domain-interacting receptors). The three Nir proteins (Nir1, Nir2, and Nir3) bind to the amino-terminal domain of PYK2 via a conserved sequence motif located in the carboxy terminus. The primary structures of Nirs reveal six putative transmembrane domains, a region homologous to phosphatidylinositol (PI) transfer protein, and an acidic domain. The Nir proteins are the human homologues of the Drosophila retinal degeneration B protein (rdgB), a protein implicated in the visual transduction pathway in flies. We demonstrate that Nirs are calcium-binding proteins that exhibit PI transfer activity in vivo. Activation of PYK2 by agents that elevate intracellular calcium or by phorbol ester induce tyrosine phosphorylation of Nirs. Moreover, PYK2 and Nirs exhibit similar expression patterns in several regions of the brain and retina. In addition, PYK2-Nir complexes are detected in lysates prepared from cultured cells or from brain tissues. Finally, the Nir1-encoding gene is located at human chromosome 17p13.1, in proximity to a locus responsible for several human retinal diseases. We propose that the Nir and rdgB proteins represent a new family of evolutionarily conserved PYK2-binding proteins that play a role in the control of calcium and phosphoinositide metabolism downstream of G-protein-coupled receptors.
Figures






References
-
- Aikawa Y, Hara H, Watanabe T. Molecular cloning and characterization of mammalian homologues of the Drosophila retinal degeneration B gene. Biochem Biophys Res Commun. 1997;236:559–564. - PubMed
-
- Astier A, Avraham H, Manie S N, Groopman J, Canty T, Avraham S, Freedman A S. The related adhesion focal tyrosine kinase is tyrosine-phosphorylated after beta1-integrin stimulation in B cells and binds to p130cas. J Biol Chem. 1997;272:228–232. - PubMed
-
- Avraham S, London R, Fu Y, Ota S, Hiregowdara D, Li J, Jiang S, Pasztor L M, White R A, Groopman J E, et al. Identification and characterization of a novel related adhesion focal tyrosine kinase (RAFTK) from megakaryocytes and brain. J Biol Chem. 1995;270:27742–27751. - PubMed
-
- Balciuniene J, Johansson K, Sandgren O, Wachtmeister L, Holmgren G, Forsman K. A gene for autosomal dominant progressive cone dystrophy (CORD5) maps to chromosome 17p12-p13. Genomics. 1995;30:281–286. - PubMed
-
- Banfi S, Borsani G, Rossi E, Bernard L, Guffanti A, Rubboli F, Marchitiello A, Giglio S, Coluccia E, Zollo M, Zuffardi O, Ballabio A. Identification and mapping of human cDNAs homologous to Drosophila mutant genes through EST database searching. Nat Genet. 1996;13:167–174. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
