The mouse Aire gene: comparative genomic sequencing, gene organization, and expression
- PMID: 10022980
- PMCID: PMC310712
The mouse Aire gene: comparative genomic sequencing, gene organization, and expression
Abstract
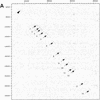
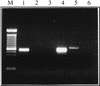
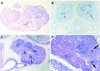
Mutations in the human AIRE gene (hAIRE) result in the development of an autoimmune disease named APECED (autoimmune polyendocrinopathy candidiasis ectodermal dystrophy; OMIM 240300). Previously, we have cloned hAIRE and shown that it codes for a putative transcription-associated factor. Here we report the cloning and characterization of Aire, the murine ortholog of hAIRE. Comparative genomic sequencing revealed that the structure of the AIRE gene is highly conserved between human and mouse. The conceptual proteins share 73% homology and feature the same typical functional domains in both species. RT-PCR analysis detected three splice variant isoforms in various mouse tissues, and interestingly one isoform was conserved in human, suggesting potential biological relevance of this product. In situ hybridization on mouse and human histological sections showed that AIRE expression pattern was mainly restricted to a few cells in the thymus, calling for a tissue-specific function of the gene product.
Figures








References
-
- Aaltonen J, Bjorses P, Sandkuijl L, Perheentupa J, Peltonen L. An autosomal locus causing autoimmune disease: Autoimmune polyglandular disease type I assigned to chromosome 21. Nat Genet. 1994;8:83–87. - PubMed
-
- Aasland R, Gibson TJ, Stewart AF. The PHD finger: Implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995;20:56–59. - PubMed
-
- Ahonen P. Autoimmune polyendocrinopathy–candidosis–ectodermal dystrophy (APECED): Autosomal recessive inheritance. Clin Gen. 1985;27:535–542. - PubMed
-
- Ahonen P, Myllarniemi S, Sipila I, Perheentupa J. Clinical variation of autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) in a series of 68 patients. N Engl J Med. 1990;322:1829–1836. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
