Evolutionary relationships of pathogenic clones of Vibrio cholerae by sequence analysis of four housekeeping genes
- PMID: 10024551
- PMCID: PMC96437
- DOI: 10.1128/IAI.67.3.1116-1124.1999
Evolutionary relationships of pathogenic clones of Vibrio cholerae by sequence analysis of four housekeeping genes
Abstract
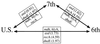
Studies of the Vibrio cholerae population, using molecular typing techniques, have shown the existence of several pathogenic clones, mainly sixth-pandemic, seventh-pandemic, and U.S. Gulf Coast clones. However, the relationship of the pathogenic clones to environmental V. cholerae isolates remains unclear. A previous study to determine the phylogeny of V. cholerae by sequencing the asd (aspartate semialdehyde dehydrogenase) gene of V. cholerae showed that the sixth-pandemic, seventh-pandemic, and U.S. Gulf Coast clones had very different asd sequences which fell into separate lineages in the V. cholerae population. As gene trees drawn from a single gene may not reflect the true topology of the population, we sequenced the mdh (malate dehydrogenase) and hlyA (hemolysin A) genes from representatives of environmental and clinical isolates of V. cholerae and found that the mdh and hlyA sequences from the three pathogenic clones were identical, except for the previously reported 11-bp deletion in hlyA in the sixth-pandemic clone. Identical sequences were obtained, despite average nucleotide differences in the mdh and hlyA genes of 1.52 and 3.25%, respectively, among all the isolates, suggesting that the three pathogenic clones are closely related. To extend these observations, segments of the recA and dnaE genes were sequenced from a selection of the pathogenic isolates, where the sequences were either identical or substantially different between the clones. The results show that the three pathogenic clones are very closely related and that there has been a high level of recombination in their evolution.
Figures



References
-
- Alm R A, Stroeher U H, Manning P A. Extracellular proteins of Vibrio cholerae: nucleotide sequence of the structural gene (hlyA) for the haemolysin of the haemolytic El Tor strain 017 and characterization of the hlyA mutation in the non-haemolytic classical strain 569B. Mol Microbiol. 1988;2:481–488. - PubMed
-
- Bastin D A, Romana L K, Reeves P R. Molecular cloning and expression in Escherichia coli K-12 of the rfb gene cluster determining the O antigen of an E. coli O111 strain. Mol Microbiol. 1991;5:2223–2231. - PubMed
-
- Berche P, Poyart C, Abachin E, Lelievre H, Vandepitte J, Dodin A, Fournier J M. The novel epidemic strain O139 is closely related to the pandemic strain O1 of Vibrio cholerae. J Infect Dis. 1994;170:701–704. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

