Molecular and evolutionary analysis of Borrelia burgdorferi 297 circular plasmid-encoded lipoproteins with OspE- and OspF-like leader peptides
- PMID: 10024606
- PMCID: PMC96492
- DOI: 10.1128/IAI.67.3.1526-1532.1999
Molecular and evolutionary analysis of Borrelia burgdorferi 297 circular plasmid-encoded lipoproteins with OspE- and OspF-like leader peptides
Abstract
We previously described two OspE and three OspF homologs in Borrelia burgdorferi 297 (D. R. Akins, S. F. Porcella, T. G. Popova, D. Shevchenko, S. I. Baker, M. Li, M. V. Norgard, and J. D. Radolf, Mol. Microbiol. 18:507-520, 1995; D. R. Akins, K. W. Bourell, M. J. Caimano, M. V. Norgard, and J. D. Radolf, J. Clin. Investig. 101:2240-2250, 1998). In this study, we characterized four additional lipoproteins with OspE/F-like leader peptides (Elps) and demonstrated that all are encoded on plasmids homologous to cp32 and cp18 from the B31 and N40 strains, respectively. Statistical analysis of sequence similarities using the binary comparison algorithm revealed that the nine lipoproteins from strain 297, as well as the OspE, OspF, and Erp proteins from the N40 and B31 strains, fall into three distinct families. Based upon the observation that these lipoproteins all contain highly conserved leader peptides, we now propose that the ancestors of each of the three families arose from gene fusion events which joined a common N terminus to unrelated proteins. Additionally, further sequence analysis of the strain 297 circular plasmids revealed that rearrangements appear to have played an important role in generating sequence diversity among the members of these three families and that recombinational events in the downstream flanking regions appear to have occurred independently of those within the lipoprotein-encoding genes. The association of hypervariable regions with genes which are differentially expressed and/or subject to immunological pressures suggests that the Lyme disease spirochete has exploited recombinatorial processes to foster its parasitic strategy and enhance its immunoevasiveness.
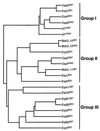
Figures


References
-
- Akins D R, Porcella S F, Popova T G, Shevchenko D, Baker S I, Li M, Norgard M V, Radolf J D. Evidence for in vivo but not in vitro expression of a Borrelia burgdorferi outer surface protein F (OspF) homolog. Mol Microbiol. 1995;18:507–520. - PubMed
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Baranton G, Postic D, Saint Girons I, Boerlin P, Piffaretti J-C, Assous M, Grimont P A D. Delineation of Borrelia burgdorferi sensu stricto, Borrelia garinii sp. nov., and group VS461 associated with Lyme borreliosis. Int J Syst Bacteriol. 1992;42:378–383. - PubMed
-
- Bergstrom S, Bundoc V G, Barbour A G. Molecular analysis of linear plasmid-encoded major surface proteins, OspA and OspB, of the Lyme disease spirochaete Borrelia burgdorferi. Mol Microbiol. 1989;3:479–486. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

