Detection and genotyping of bovine diarrhea virus by reverse transcription-polymerase chain amplification of the 5' untranslated region
- PMID: 10028170
- PMCID: PMC7117503
- DOI: 10.1016/s0378-1135(98)00267-3
Detection and genotyping of bovine diarrhea virus by reverse transcription-polymerase chain amplification of the 5' untranslated region
Abstract
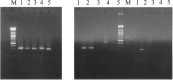
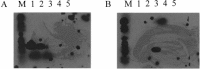
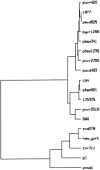
A reverse-transcription polymerase chain reaction (RT-PCR) was developed to differentiate the bovine diarrhea virus (BVDV) from other pestiviruses, and to determine the genotype of the BVDV isolates. For this purpose, primer pairs were selected in the 5' untranslated region (5'UTR). The primers BE and B2 were located in highly conserved regions and were pestivirus-specific. Two primer pairs named B3B4 and B5B6 were specific of BVDV genotypes I and II, respectively. With this technique, an amplification product of the expected size was obtained with either the B3B4 or the B5B6 primer pairs for the 107 BVDV isolates tested but not for BDV or CSFV. For some isolates that were grouped in the genotype II, sequence analysis of the PCR fragments confirmed their classification into this genotype.
Figures

Similar articles
-
Molecular analysis of bovine viral diarrhoea virus isolates from South Africa.Onderstepoort J Vet Res. 2003 Dec;70(4):273-9. doi: 10.4102/ojvr.v70i4.292. Onderstepoort J Vet Res. 2003. PMID: 14971730
-
Genetic typing of bovine pestiviruses from England and Wales.Vet Microbiol. 1999 Sep 29;69(4):227-37. doi: 10.1016/s0378-1135(99)00111-x. Vet Microbiol. 1999. PMID: 10535769
-
Genotyping and phylogenetic analysis of bovine viral diarrhea virus isolates from BVDV infected alpacas in North America.Vet Microbiol. 2009 May 12;136(3-4):209-16. doi: 10.1016/j.vetmic.2008.10.029. Epub 2008 Nov 5. Vet Microbiol. 2009. PMID: 19059738
-
BVDV genotypes and biotypes: practical implications for diagnosis and control.Biologicals. 2003 Jun;31(2):127-31. doi: 10.1016/s1045-1056(03)00028-9. Biologicals. 2003. PMID: 12770544 Review. No abstract available.
-
Molecular biology of bovine viral diarrhea virus and its interactions with the host.Vet Clin North Am Food Anim Pract. 1995 Nov;11(3):393-423. doi: 10.1016/s0749-0720(15)30459-x. Vet Clin North Am Food Anim Pract. 1995. PMID: 8581855 Free PMC article. Review.
Cited by
-
Seroprevalence of border disease virus and other pestiviruses in sheep in Algeria and associated risk factors.BMC Vet Res. 2018 Nov 12;14(1):339. doi: 10.1186/s12917-018-1666-y. BMC Vet Res. 2018. PMID: 30419908 Free PMC article.
-
Comparative analysis on the 5'-untranslated region of bovine viral diarrhea virus isolated in Korea.Res Vet Sci. 2004 Apr;76(2):157-63. doi: 10.1016/j.rvsc.2003.10.003. Res Vet Sci. 2004. PMID: 14672860 Free PMC article.
-
Persistent bovine viral diarrhea virus (BVDV) infection in cattle herds.Iran J Vet Res. 2017 Summer;18(3):154-163. Iran J Vet Res. 2017. PMID: 29163643 Free PMC article. Review.
-
Persistent BVD virus infections in offspring from imported heifers.Trop Anim Health Prod. 2019 Feb;51(2):297-302. doi: 10.1007/s11250-018-1685-5. Epub 2018 Aug 18. Trop Anim Health Prod. 2019. PMID: 30121755
-
Epidemiology and genetic characterization of BVDV, BHV-1, BHV-4, BHV-5 and Brucella spp. infections in cattle in Turkey.J Vet Med Sci. 2015 Nov;77(11):1371-7. doi: 10.1292/jvms.14-0657. Epub 2015 Jun 21. J Vet Med Sci. 2015. PMID: 26096964 Free PMC article.
References
-
- Becher P., König M., Paton D.J., Thiel H.-J. Further characterization of border disease virus isolates: Evidence for the presence of more than three species within the genus pestivirus. Virology. 1995;209:200–206. - PubMed
-
- Becher P., Orlich M., Shannon A.D., Horner G., König M., Thiel H.-J. Phylogenic analysis of pestiviruses from domestic and wild ruminants. J. Gen. Virol. 1997;78:1357–1366. - PubMed
-
- Broes A., Wellemans G., Dheedene J. Syndrome hémorragique chez des bovins infectés par le virus de la diarrhée bovine (BVD/MD) Ann. Méd. Vét. 1992;137:33–38.
-
- Brownlie, J., 1991. The pathways for bovine virus diarrhoea virus biotypes in the pathogenesis of disease. Arch. Virol. (Suppl 3), 79–96 - PubMed
-
- Canal C.W., Hotzel I., de Almeida L.L., Roehe P.M., Masuda A. Differentiation of classical swine fever virus from ruminant pestiviruses by reverse transcription and polymerase chain reaction. Vet. Microbiol. 1996;48:373–379. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

