LYVE-1, a new homologue of the CD44 glycoprotein, is a lymph-specific receptor for hyaluronan
- PMID: 10037799
- PMCID: PMC2132933
- DOI: 10.1083/jcb.144.4.789
LYVE-1, a new homologue of the CD44 glycoprotein, is a lymph-specific receptor for hyaluronan
Abstract
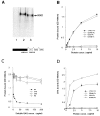
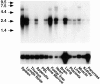
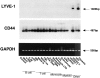
The extracellular matrix glycosaminoglycan hyaluronan (HA) is an abundant component of skin and mesenchymal tissues where it facilitates cell migration during wound healing, inflammation, and embryonic morphogenesis. Both during normal tissue homeostasis and particularly after tissue injury, HA is mobilized from these sites through lymphatic vessels to the lymph nodes where it is degraded before entering the circulation for rapid uptake by the liver. Currently, however, the identities of HA binding molecules which control this pathway are unknown. Here we describe the first such molecule, LYVE-1, which we have identified as a major receptor for HA on the lymph vessel wall. The deduced amino acid sequence of LYVE-1 predicts a 322-residue type I integral membrane polypeptide 41% similar to the CD44 HA receptor with a 212-residue extracellular domain containing a single Link module the prototypic HA binding domain of the Link protein superfamily. Like CD44, the LYVE-1 molecule binds both soluble and immobilized HA. However, unlike CD44, the LYVE-1 molecule colocalizes with HA on the luminal face of the lymph vessel wall and is completely absent from blood vessels. Hence, LYVE-1 is the first lymph-specific HA receptor to be characterized and is a uniquely powerful marker for lymph vessels themselves.
Figures









References
-
- Aruffo A, Stamenkovic I, Melnick M, Underhill CB, Seed B. CD44 is the principal cell surface receptor for hyaluronate. Cell. 1990;61:1303–1313. - PubMed
-
- Bajorath J, Greenfield B, Munro SB, Day AJ, Aruffo A. Identification of CD44 residues important for hyaluronan binding and delineation of the binding site. J Biol Chem. 1998;273:338–343. - PubMed
-
- Banerji S, Day AJ, Kahmann JD, Jackson DG. Characterization of a functional hyaluronan-binding domain from the human CD44 molecule expressed in Escherichia coli. . Protein Exp Purification. 1998;14:371–381. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

