The Caenorhabditis elegans gene ham-2 links Hox patterning to migration of the HSN motor neuron
- PMID: 10049362
- PMCID: PMC316472
- DOI: 10.1101/gad.13.4.472
The Caenorhabditis elegans gene ham-2 links Hox patterning to migration of the HSN motor neuron
Abstract
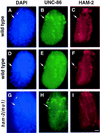
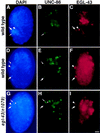
The Caenorhabditis elegans HSN motor neurons permit genetic analysis of neuronal development at single-cell resolution. The egl-5 Hox gene, which patterns the posterior of the embryo, is required for both early (embryonic) and late (larval) development of the HSN. Here we show that ham-2 encodes a zinc finger protein that acts downstream of egl-5 to direct HSN cell migration, an early differentiation event. We also demonstrate that the EGL-43 zinc finger protein, also required for HSN migration, is expressed in the HSN specifically during its migration. In an egl-5 mutant background, the HSN still expresses EGL-43, but expression is no longer down-regulated at the end of the cell's migration. Finally, we find a new role in early HSN differentiation for UNC-86, a POU homeodomain transcription factor shown previously to act downstream of egl-5 in the regulation of late HSN differentiation. In an unc-86; ham-2 double mutant the HSNs are defective in EGL-43 down-regulation, an egl-5-like phenotype that is absent in either single mutant. Thus, in the HSN, a Hox gene, egl-5, regulates cell fate by activating the transcription of genes encoding the transcription factors HAM-2 and UNC-86 that in turn individually control some differentiation events and combinatorially affect others.
Figures





References
-
- Bang AG, Goulding MD. Regulation of vertebrate neural cell fate by transcription factors. Curr Opin in Neurobiol. 1996;6:25–32. - PubMed
-
- Basson M, Horvitz HR. The Caenorhabditis elegans gene sem-4 controls neuronal and mesodermal cell development and encodes a zinc finger protein. Genes & Dev. 1996;10:1953–1965. - PubMed
-
- Baumeister R, Liu Y, Ruvkun G. Lineage-specific regulators couple cell lineage asymmetry to the transcription of the Caenorhabditis elegans POU gene unc-86 during neurogenesis. Genes & Dev. 1996;10:1395–1410. - PubMed
-
- Berg JM, Shi Y. The galvanization of biology: A growing appreciation for the roles of zinc. Science. 1996;271:1081–1085. - PubMed
-
- Blumenthal T. Trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet. 1995;11:132–136. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
