Role of the Bicoid-related homeodomain factor Pitx1 in specifying hindlimb morphogenesis and pituitary development
- PMID: 10049363
- PMCID: PMC316471
- DOI: 10.1101/gad.13.4.484
Role of the Bicoid-related homeodomain factor Pitx1 in specifying hindlimb morphogenesis and pituitary development
Abstract
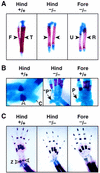
Pitx1 is a Bicoid-related homeodomain factor that exhibits preferential expression in the hindlimb, as well as expression in the developing anterior pituitary gland and first branchial arch. Here, we report that Pitx1 gene-deleted mice exhibit striking abnormalities in morphogenesis and growth of the hindlimb, resulting in a limb that exhibits structural changes in tibia and fibula as well as patterning alterations in patella and proximal tarsus, to more closely resemble the corresponding forelimb structures. Deletion of the Pitx1 locus results in decreased distal expression of the hindlimb-specific marker, the T-box factor, Tbx4. On the basis of similar expression patterns in chick, targeted misexpression of chick Pitx1 in the developing wing bud causes the resulting limb to assume altered digit number and morphogenesis, with Tbx4 induction. We hypothesize that Pitx1 serves to critically modulate morphogenesis, growth, and potential patterning of a specific hindlimb region, serving as a component of the morphological and growth distinctions in forelimb and hindlimb identity. Pitx1 gene-deleted mice also exhibit reciprocal abnormalities of two ventral and one dorsal anterior pituitary cell types, presumably on the basis of its synergistic functions with other transcription factors, and defects in the derivatives of the first branchial arch, including cleft palate, suggesting a proliferative defect in these organs analogous to that observed in the hindlimb.
Figures






References
-
- Acampora D, Mazan S, Lallemand Y, Avantaggiato V, Maury M, Simeone A, Brûlet P. Forebrain and midbrain regions are deleted in Otx2-/- mutants due to a defective anterior neuroectoderm specification during gastrulation. Development. 1995;121:3279–3290. - PubMed
-
- Acampora D, Mazan S, Avantaggiato V, Barone P, Tuorto F, Lallemand Y, Brûlet P, Simeone A. Epilepsy and brain abnormalities in mice lacking the Otx1 gene. Nat Genet. 1996;14:218–222. - PubMed
-
- Acampora D, Mazan S, Tuorto F, Avantaggiato V, Tremblay JJ, Lazzaro D, di Carlo A, Mariano A, Macchia PE, Corte G, Macchia V, Drouin J, Brulet P, Simeone A. Transient dwarfism and hypogonadism in mice lacking Otx-1 reveal prepubescent stage-specific control of pituitary levels of Gh, FSH and LH. Development. 1998;125:1229–1239. - PubMed
-
- Ang SL, Jin O, Rhinn M, Daigle N, Stevenson L, Rossant J. A targeted mouse Otx2 mutation leads to severe defects in gastrulation and formation of axial mesoderm and to deletion of rostral brain. Development. 1996;122:243–252. - PubMed
-
- Bermingham JR, Jr, Scherer SS, O’Connell S, Arroyo E, Kalla KA, Powell FL, Rosenfeld MG. Tst-1/Oct-6/SCIP regulates a unique step in peripheral myelination and is required for normal respiration. Genes & Dev. 1996;10:1751–1762. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
