Peptidoglycan-hydrolyzing activity of the FlgJ protein, essential for flagellar rod formation in Salmonella typhimurium
- PMID: 10049388
- PMCID: PMC93546
- DOI: 10.1128/JB.181.5.1555-1561.1999
Peptidoglycan-hydrolyzing activity of the FlgJ protein, essential for flagellar rod formation in Salmonella typhimurium
Abstract
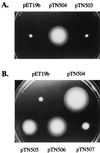
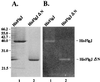
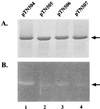
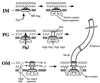
Because the rod structure of the flagellar basal body crosses the inner membrane, the periplasmic space, and the outer membrane, its formation must involve hydrolysis of the peptidoglycan layer. So far, more than 10 genes have been shown to be required for rod formation in Salmonella typhimurium. Some of them encode the component proteins of the rod structure, and most of the remaining genes are believed to encode proteins involved in the export process of the component proteins. Although FlgJ has also been known to be involved in rod formation, its exact role has not been understood. Recently, it was suggested that the C-terminal half of the FlgJ protein has homology to the active center of some muramidase enzymes from gram-positive bacteria. In this study, we showed that the purified FlgJ protein from S. typhimurium has a peptidoglycan-hydrolyzing activity and that this activity is localized in its C-terminal half. Through oligonucleotide-directed mutagenesis, we constructed flgJ mutants with amino acid substitutions in the putative active center of the muramidase. The resulting mutants produced FlgJ proteins with reduced enzymatic activity and showed poor motility. These results indicate that the muramidase activity of FlgJ is essential for flagellar formation. Immunoblotting analysis with the fractionated cell extracts revealed that FlgJ is exported to the periplasmic space, where the peptidoglycan layer is localized. On the basis of these results, we conclude that FlgJ is the flagellum-specific muramidase which hydrolyzes the peptidoglycan layer to assemble the rod structure in the periplasmic space.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

