Small antibody-like proteins with prescribed ligand specificities derived from the lipocalin fold
- PMID: 10051566
- PMCID: PMC26708
- DOI: 10.1073/pnas.96.5.1898
Small antibody-like proteins with prescribed ligand specificities derived from the lipocalin fold
Abstract
We demonstrate that the ligand pocket of a lipocalin from Pieris brassicae, the bilin-binding protein (BBP), can be reshaped by combinatorial protein design such that it recognizes fluorescein, an established immunological hapten. For this purpose 16 residues at the center of the binding site, which is formed by four loops on top of an eight-stranded beta-barrel, were subjected to random mutagenesis. Fluorescein-binding BBP variants were then selected from the mutant library by bacterial phage display. Three variants were identified that complex fluorescein with high affinity, exhibiting dissociation constants as low as 35.2 nM. Notably, one of these variants effects almost complete quenching of the ligand fluorescence, similarly as an anti-fluorescein antibody. Detailed ligand-binding studies and site-directed mutagenesis experiments indicated (i) that the molecular recognition of fluorescein is specific and (ii) that charged residues at the center of the pocket are responsible for tight complex formation. Sequence comparison of the BBP variants directed against fluorescein with the wild-type protein and with further variants that were selected against several other ligands revealed that all of the randomized amino acid positions are variable. Hence, a lipocalin can be used for generating molecular pockets with a diversity of shapes. We term this class of engineered proteins "anticalins." Their one-domain scaffold makes them a promising alternative to antibodies to create a stable receptor protein for a ligand of choice.
Figures
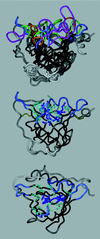

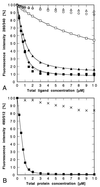
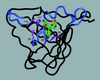
References
-
- Skerra A. Curr Opin Immunol. 1993;5:256–262. - PubMed
-
- Winter G, Milstein C. Nature (London) 1991;349:293–299. - PubMed
-
- Hoogenboom H R. Trends Biotechnol. 1997;15:62–70. - PubMed
-
- Huber R, Schneider M, Mayr I, Müller R, Deutzmann R, Suter F, Zuber H, Falk H, Kayser H. J Mol Biol. 1987;198:499–513. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

