An Escherichia coli strain with all chromosomal rRNA operons inactivated: complete exchange of rRNA genes between bacteria
- PMID: 10051579
- PMCID: PMC26721
- DOI: 10.1073/pnas.96.5.1971
An Escherichia coli strain with all chromosomal rRNA operons inactivated: complete exchange of rRNA genes between bacteria
Abstract
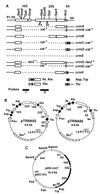
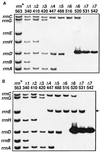
Current global phylogenies are built predominantly on rRNA sequences. However, an experimental system for studying the evolution of rRNA is not readily available, mainly because the rRNA genes are highly repeated in most experimental organisms. We have constructed an Escherichia coli strain in which all seven chromosomal rRNA operons are inactivated by deletions spanning the 16S and 23S coding regions. A single E. coli rRNA operon carried by a multicopy plasmid supplies 16S and 23S rRNA to the cell. By using this strain we have succeeded in creating microorganisms that contain only a foreign rRNA operon derived from either Salmonella typhimurium or Proteus vulgaris, microorganisms that have diverged from E. coli about 120-350 million years ago. We also were able to replace the E. coli rRNA operon with an E. coli/yeast hybrid one in which the GTPase center of E. coli 23S rRNA had been substituted by the corresponding domain from Saccharomyces cerevisiae. These results suggest that, contrary to common belief, coevolution of rRNA with many other components in the translational machinery may not completely preclude the horizontal transfer of rRNA genes.
Figures



Comment on
-
Engineering of bacterial ribosomes: replacement of all seven Escherichia coli rRNA operons by a single plasmid-encoded operon.Proc Natl Acad Sci U S A. 1999 Mar 2;96(5):1820-2. doi: 10.1073/pnas.96.5.1820. Proc Natl Acad Sci U S A. 1999. PMID: 10051551 Free PMC article. Review. No abstract available.
References
-
- Gesteland R F, Atkins J F, editors. The RNA World. Plainview, NY: Cold Spring Harbor Lab. Press; 1993.
-
- Noller H, Hoffarth V, Zimniak L. Science. 1992;256:1416–1419. - PubMed
-
- Nitta I, Kamada Y, Noda H, Ueda T, Watanabe K. Science. 1998;281:666–669. - PubMed
-
- Ludwig W, Strunk O, Klugbauer S, Klugbauer N, Weizenegger M, Neumaier J, Bachleitner M, Schleifer K H. Electrophoresis. 1998;19:554–568. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

