Ligand-dependent activation of transcription in vitro by retinoic acid receptor alpha/retinoid X receptor alpha heterodimers that mimics transactivation by retinoids in vivo
- PMID: 10051583
- PMCID: PMC26725
- DOI: 10.1073/pnas.96.5.1995
Ligand-dependent activation of transcription in vitro by retinoic acid receptor alpha/retinoid X receptor alpha heterodimers that mimics transactivation by retinoids in vivo
Abstract
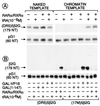
All-trans and 9-cis retinoic acids (RA) signals are transduced by retinoic acid receptor/retinoid X receptor (RAR/RXR) heterodimers that act as functional units controlling the transcription of RA-responsive genes. With the aim of elucidating the underlying molecular mechanisms, we have developed an in vitro transcription system using a chromatin template made up of a minimal promoter and a direct repeat with 5-spacing-based RA response element. RARalpha and RXRalpha were expressed in and purified from baculovirus-infected Sf9 cells, and transcription was carried out by using naked DNA or chromatin templates. Transcription from naked templates was not affected by the presence of RA and/or RAR/RXR heterodimers. In contrast, very little transcription occurred from chromatin templates in the absence of RA or RAR/RXR heterodimers whereas their addition resulted in a dosage-dependent stimulation of transcription that never exceeded that occurring on naked DNA templates. Most importantly, the addition of synthetic agonistic or antagonistic retinoids to the chromatin transcription system mimicked their stimulatory or inhibitory action in vivo, and activation by a RXR-specific retinoid was subordinated to the binding of an agonist ligand to the RAR partner. Moreover, the addition of the p300 coactivator generated a synergistic enhancement of transcription. Thus, the dissection of this transcription system ultimately should lead to the elucidation of the molecular mechanisms by which RAR/RXR heterodimers control transcription in a ligand-dependent manner.
Figures

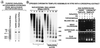

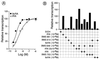

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

