Copper binding to the prion protein: structural implications of four identical cooperative binding sites
- PMID: 10051591
- PMCID: PMC26733
- DOI: 10.1073/pnas.96.5.2042
Copper binding to the prion protein: structural implications of four identical cooperative binding sites
Abstract
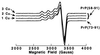
Evidence is growing to support a functional role for the prion protein (PrP) in copper metabolism. Copper ions appear to bind to the protein in a highly conserved octapeptide repeat region (sequence PHGGGWGQ) near the N terminus. To delineate the site and mode of binding of Cu(II) to the PrP, the copper-binding properties of peptides of varying lengths corresponding to 2-, 3-, and 4-octarepeat sequences have been probed by using various spectroscopic techniques. A two-octarepeat peptide binds a single Cu(II) ion with Kd approximately 6 microM whereas a four-octarepeat peptide cooperatively binds four Cu(II) ions. Circular dichroism spectra indicate a distinctive structuring of the octarepeat region on Cu(II) binding. Visible absorption, visible circular dichroism, and electron spin resonance spectra suggest that the coordination sphere of the copper is identical for 2, 3, or 4 octarepeats, consisting of a square-planar geometry with three nitrogen ligands and one oxygen ligand. Consistent with the pH dependence of Cu(II) binding, proton NMR spectroscopy indicates that the histidine residues in each octarepeat are coordinated to the Cu(II) ion. Our working model for the structure of the complex shows the histidine residues in successive octarepeats bridged between two copper ions, with both the Nepsilon2 and Ndelta1 imidazole nitrogen of each histidine residue coordinated and the remaining coordination sites occupied by a backbone amide nitrogen and a water molecule. This arrangement accounts for the cooperative nature of complex formation and for the apparent evolutionary requirement for four octarepeats in the PrP.
Figures




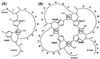
References
-
- Prusiner S B. Science. 1997;278:245–251. - PubMed
-
- Chazot G, Broussolle E, Lapras C, Blattler T, Aguzzi A, Kopp N. Lancet. 1996;347:1181. - PubMed
-
- Will R G, Ironside J W, Zeidler M, Cousens S N, Estibeiro K, Alperovitch A, Poser S, Pocchiari M, Hofman A, Smith P G. Lancet. 1996;347:921–925. - PubMed
-
- Prusiner S B. Science. 1982;216:136–144. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

