Human deafness dystonia syndrome is a mitochondrial disease
- PMID: 10051608
- PMCID: PMC26750
- DOI: 10.1073/pnas.96.5.2141
Human deafness dystonia syndrome is a mitochondrial disease
Abstract
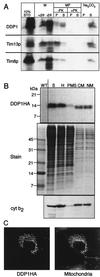
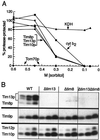
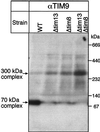
The human deafness dystonia syndrome results from the mutation of a protein (DDP) of unknown function. We show now that DDP is a mitochondrial protein and similar to five small proteins (Tim8p, Tim9p, Tim10p, Tim12p, and Tim13p) of the yeast mitochondrial intermembrane space. Tim9p, Tim10p, and Tim12p mediate the import of metabolite transporters from the cytoplasm into the mitochondrial inner membrane and interact structurally and functionally with Tim8p and Tim13p. DDP is most similar to Tim8p. Tim8p exists as a soluble 70-kDa complex with Tim13p and Tim9p, and deletion of Tim8p is synthetically lethal with a conditional mutation in Tim10p. The deafness dystonia syndrome thus is a novel type of mitochondrial disease that probably is caused by a defective mitochondrial protein-import system.
Figures


Comment in
-
Mitochondria and dystonia: the movement disorder connection?Proc Natl Acad Sci U S A. 1999 Mar 2;96(5):1817-9. doi: 10.1073/pnas.96.5.1817. Proc Natl Acad Sci U S A. 1999. PMID: 10051550 Free PMC article. No abstract available.
References
-
- Ryan K R, Jensen R E. Cell. 1995;83:517–519. - PubMed
-
- Schatz G, Dobberstein B. Science. 1996;271:1519–1526. - PubMed
-
- Neupert W. Annu Rev Biochem. 1997;66:863–917. - PubMed
-
- Pfanner N. Curr Biol. 1998;8:R262–R265. - PubMed
-
- Koehler C M, Jarosch E, Tokatlidis K, Schmid K, Schweyen R J, Schatz G. Science. 1998;279:369–373. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

