Precise packaging of the three genomic segments of the double-stranded-RNA bacteriophage phi6
- PMID: 10066834
- PMCID: PMC98960
- DOI: 10.1128/MMBR.63.1.149-160.1999
Precise packaging of the three genomic segments of the double-stranded-RNA bacteriophage phi6
Abstract
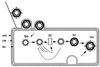
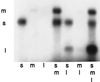
Bacteriophage phi6 has a genome of three segments of double-stranded RNA. Each virus particle contains one each of the three segments. Packaging is effected by the acquisition, in a serially dependent manner, of the plus strands of the genomic segments into empty procapsids. The empty procapsids are compressed in shape and expand during packaging. The packaging program involves discrete steps that are determined by the amount of RNA inside the procapsid. The steps involve the exposure and concealment of binding sites on the outer surface of the procapsid for the plus strands of the three genomic segments. The plus strand of segment S can be packaged alone, while packaging of the plus strand of segment M depends upon prior packaging of S. Packaging of the plus strand of L depends upon the prior packaging of M. Minus-strand synthesis begins when the particle has a full complement of plus strands. Plus-strand synthesis commences upon the completion of minus-strand synthesis. All of the reactions of packaging, minus-strand synthesis, and plus-strand synthesis can be accomplished in vitro with isolated procapsids. Live-virus constructions that are in accord with the model have been prepared. Mutant virus with changes in the packaging program have been isolated and analyzed.
Figures









References
-
- Black L W. DNA packaging in dsDNA bacteriophages. In: Calendar R, editor. The bacteriophages. Vol. 2. New York, N.Y: Plenum Publishing Corp.; 1988. pp. 321–373.
-
- Caldentey J, Bamford D H. The lytic enzyme of the Pseudomonas phage φ6. Purification and biochemical characterization. Biochim Biopys Acta. 1992;1159:44–50. - PubMed
-
- Casini G, Qiao X, Mindich L. Reconstitution of active replicase in procapsids of the segmented dsRNA bacteriophage φ6. Virology. 1994;204:251–253. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

