Interaction of the Doa4 deubiquitinating enzyme with the yeast 26S proteasome
- PMID: 10069815
- PMCID: PMC25199
- DOI: 10.1091/mbc.10.3.741
Interaction of the Doa4 deubiquitinating enzyme with the yeast 26S proteasome
Abstract
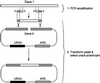
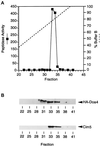
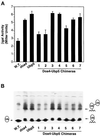
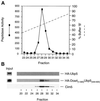
e Saccharomyces cerevisiae Doa4 deubiquitinating enzyme is required for the rapid degradation of protein substrates of the ubiquitin-proteasome pathway. Previous work suggested that Doa4 functions late in the pathway, possibly by deubiquitinating (poly)-ubiquitin-substrate intermediates associated with the 26S proteasome. We now provide evidence for physical and functional interaction between Doa4 and the proteasome. Genetic interaction is indicated by the mutual enhancement of defects associated with a deletion of DOA4 or a proteasome mutation when the two mutations are combined. Physical association of Doa4 and the proteasome was investigated with a new yeast 26S proteasome purification procedure, by which we find that a sizeable fraction of Doa4 copurifies with the protease. Another yeast deubiquitinating enzyme, Ubp5, which is related in sequence to Doa4 but cannot substitute for it even when overproduced, does not associate with the proteasome. DOA4-UBP5 chimeras were made by a novel PCR/yeast recombination method and used to identify an N-terminal 310-residue domain of Doa4 that, when appended to the catalytic domain of Ubp5, conferred Doa4 function, consistent with Ubp enzymes having a modular architecture. Unlike Ubp5, a functional Doa4-Ubp5 chimera associates with the proteasome, suggesting that proteasome binding is important for Doa4 function. Together, these data support a model in which Doa4 promotes proteolysis through removal of ubiquitin from proteolytic intermediates on the proteasome before or after initiation of substrate breakdown.
Figures






References
-
- Akopian TN, Kisselev AF, Goldberg AL. Processive degradation of proteins and other catalytic properties of the proteasome from Thermoplasma acidophilum. J Biol Chem. 1997;272:1791–1798. - PubMed
-
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1989.
-
- Baker RT, Tobias JW, Varshavsky A. Ubiquitin-specific proteases of Saccharomyces cerevisiae. Cloning of UBP2 and UBP3, and functional analysis of the UBP gene family. J Biol Chem. 1992;267:23364–23375. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

