Short DNA fragments without sequence similarity are initiation sites for replication in the chromosome of the yeast Yarrowia lipolytica
- PMID: 10069816
- PMCID: PMC25200
- DOI: 10.1091/mbc.10.3.757
Short DNA fragments without sequence similarity are initiation sites for replication in the chromosome of the yeast Yarrowia lipolytica
Abstract
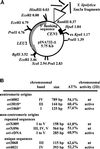
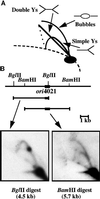
We have previously shown that both a centromere (CEN) and a replication origin are necessary for plasmid maintenance in the yeast Yarrowia lipolytica (). Because of this requirement, only a small number of centromere-proximal replication origins have been isolated from Yarrowia. We used a CEN-based plasmid to obtain noncentromeric origins, and several new fragments, some unique and some repetitive sequences, were isolated. Some of them were analyzed by two-dimensional gel electrophoresis and correspond to actual sites of initiation (ORI) on the chromosome. We observed that a 125-bp fragment is sufficient for a functional ORI on plasmid, and that chromosomal origins moved to ectopic sites on the chromosome continue to act as initiation sites. These Yarrowia origins share an 8-bp motif, which is not essential for origin function on plasmids. The Yarrowia origins do not display any obvious common structural features, like bent DNA or DNA unwinding elements, generally present at or near eukaryotic replication origins. Y. lipolytica origins thus share features of those in the unicellular Saccharomyces cerevisiae and in multicellular eukaryotes: they are discrete and short genetic elements without sequence similarity.
Figures



Similar articles
-
Only centromeres can supply the partition system required for ARS function in the yeast Yarrowia lipolytica.J Mol Biol. 2001 Jan 12;305(2):203-17. doi: 10.1006/jmbi.2000.4300. J Mol Biol. 2001. PMID: 11124900
-
An origin of replication and a centromere are both needed to establish a replicative plasmid in the yeast Yarrowia lipolytica.Mol Cell Biol. 1997 Apr;17(4):1995-2004. doi: 10.1128/MCB.17.4.1995. Mol Cell Biol. 1997. PMID: 9121447 Free PMC article.
-
Colocalization of centromeric and replicative functions on autonomously replicating sequences isolated from the yeast Yarrowia lipolytica.Proc Natl Acad Sci U S A. 1993 Jun 1;90(11):4912-6. doi: 10.1073/pnas.90.11.4912. Proc Natl Acad Sci U S A. 1993. PMID: 8506336 Free PMC article.
-
Mapping replication origins in yeast chromosomes.Bioessays. 1991 Jul;13(7):317-22. doi: 10.1002/bies.950130702. Bioessays. 1991. PMID: 1759974 Review.
-
Structure, replication efficiency and fragility of yeast ARS elements.Res Microbiol. 2012 May;163(4):243-53. doi: 10.1016/j.resmic.2012.03.003. Epub 2012 Mar 28. Res Microbiol. 2012. PMID: 22504206 Review.
Cited by
-
Establishment of a near-contiguous genome sequence of the citric acid producing yeast Yarrowia lipolytica DSM 3286 with resolution of rDNA clusters and telomeres.NAR Genom Bioinform. 2021 Oct 14;3(4):lqab085. doi: 10.1093/nargab/lqab085. eCollection 2021 Dec. NAR Genom Bioinform. 2021. PMID: 34661101 Free PMC article.
-
Choosing a suitable method for the identification of replication origins in microbial genomes.Front Microbiol. 2015 Sep 30;6:1049. doi: 10.3389/fmicb.2015.01049. eCollection 2015. Front Microbiol. 2015. PMID: 26483774 Free PMC article. Review.
-
DNA elements modulating the KARS12 chromosomal replicator in Kluyveromyces lactis.Mol Genet Genomics. 2007 Mar;277(3):287-99. doi: 10.1007/s00438-006-0188-7. Epub 2006 Nov 29. Mol Genet Genomics. 2007. PMID: 17136349
-
DNA bending in the replication zone of the C3 DNA puff amplicon of Rhynchosciara americana (Diptera: Sciaridae).Mol Biol Rep. 2006 Mar;33(1):71-82. doi: 10.1007/s11033-006-0009-4. Mol Biol Rep. 2006. PMID: 16636920
References
-
- Amati BB, Gasser SM. Chromosomal ARS and CEN elements bind specifically to the yeast nuclear scaffold. Cell. 1988;54:967–978. - PubMed
-
- Barth G, Gaillardin C. Yarrowia lipolytica. In: Wolf K, editor. Nonconventional Yeasts in Biotechnology: A Handbook. Heidelberg: Springer-Verlag; 1996. pp. 313–388.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

