A single limit dextrinase gene is expressed both in the developing endosperm and in germinated grains of barley
- PMID: 10069825
- PMCID: PMC32101
- DOI: 10.1104/pp.119.3.859
A single limit dextrinase gene is expressed both in the developing endosperm and in germinated grains of barley
Abstract
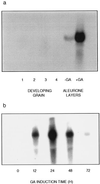
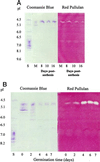
The single gene encoding limit dextrinase (pullulan 6-glucanohydrolase; EC 3.2.1.41) in barley (Hordeum vulgare) has 26 introns that range in size from 93 to 822 base pairs. The mature polypeptide encoded by the gene has 884 amino acid residues and a calculated molecular mass of 97,417 D. Limit dextrinase mRNA is abundant in gibberellic acid-treated aleurone layers and in germinated grain. Gibberellic acid response elements were found in the promoter region of the gene. These observations suggest that the enzyme participates in starch hydrolysis during endosperm mobilization in germinated grain. The mRNA encoding the enzyme is present at lower levels in the developing endosperm of immature grain, a location consistent with a role for limit dextrinase in starch synthesis. Enzyme activity was also detected in developing grain. The limit dextrinase has a presequence typical of transit peptides that target nascent polypeptides to amyloplasts, but this would not be expected to direct secretion of the mature enzyme from aleurone cells in germinated grain. It remains to be discovered how the enzyme is released from the aleurone and whether another enzyme, possibly of the isoamylase group, might be equally important for starch hydrolysis in germinated grain.
Figures



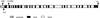



References
-
- Aman P, Hesselman K, Tilly A-C. The variation in chemical composition of Swedish barleys. J Cereal Sci. 1985;3:73–77.
-
- Amemura A, Chakraborty R, Fujita M, Noumi T, Futai M. Cloning and nucleotide sequence of the isoamylase gene from Pseudomonas amyloderamosa SB-15. J Biol Chem. 1988;263:9271–9275. - PubMed
-
- Ball S, Guan H-P, James M, Myers A, Keeling P, Mouille G, Buleon A, Colonna P, Preiss J. From glycogen to amylopectin: a model for the biogenesis of the plant starch granule. Cell. 1996;86:349–352. - PubMed
-
- Banik M, Garrett TPJ, Fincher GB. Molecular cloning of cDNAs encoding (1→4)-β-xylan endohydrolases from the aleurone layer of germinated barley (Hordeum vulgare) Plant Mol Biol. 1996;31:1163–1172. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

