The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast
- PMID: 10072382
- PMCID: PMC316509
- DOI: 10.1101/gad.13.5.532
The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast
Abstract
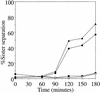
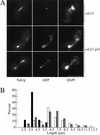
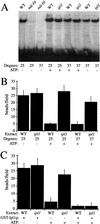
Chromosome segregation depends on kinetochores, the structures that mediate chromosome attachment to the mitotic spindle. We isolated mutants in IPL1, which encodes a protein kinase, in a screen for budding yeast mutants that have defects in sister chromatid separation and segregation. Cytological tests show that ipl1 mutants can separate sister chromatids but are defective in chromosome segregation. Kinetochores assembled in extracts from ipl1 mutants show altered binding to microtubules. Ipl1p phosphorylates the kinetochore component Ndc10p in vitro and we propose that Ipl1p regulates kinetochore function via Ndc10p phosphorylation. Ipl1p localizes to the mitotic spindle and its levels are regulated during the cell cycle. This pattern of localization and regulation is similar to that of Ipl1p homologs in higher eukaryotes, such as the human aurora2 protein. Because aurora2 has been implicated in oncogenesis, defects in kinetochore function may contribute to genetic instability in human tumors.
Figures





References
-
- Axton JM, Dombradi V, Cohen PT, Glover DM. One of the protein phosphatase 1 isoenzymes in Drosophila is for mitosis. Cell. 1990;63:33–46. - PubMed
-
- Biggins S, Murray A. Sister chromatid cohesion in mitosis. Curr Opin Cell Biol. 1998;10:769–775. - PubMed
-
- Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
