Interaction of the U1 snRNP with nonconserved intronic sequences affects 5' splice site selection
- PMID: 10072385
- PMCID: PMC316504
- DOI: 10.1101/gad.13.5.569
Interaction of the U1 snRNP with nonconserved intronic sequences affects 5' splice site selection
Abstract
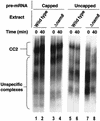
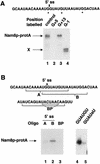
Intron definition and splice site selection occur at an early stage during assembly of the spliceosome, the complex mediating pre-mRNA splicing. Association of U1 snRNP with the pre-mRNA is required for these early steps. We report here that the yeast U1 snRNP-specific protein Nam8p is a component of the commitment complexes, the first stable complexes assembled on pre-mRNA. In vitro and in vivo, Nam8p becomes indispensable for efficient 5' splice site recognition when this process is impaired as a result of the presence of noncanonical 5' splice sites or the absence of a cap structure. Nam8p stabilizes commitment complexes in the latter conditions. Consistent with this, Nam8p interacts with the pre-mRNA downstream of the 5' splice site, in a region of nonconserved sequence. Substitutions in this region affect splicing efficiency and alternative splice site choice in a Nam8p-dependent manner. Therefore, Nam8p is involved in a novel mechanism by which a snRNP component can affect splice site choice and regulate intron removal through its interaction with a nonconserved sequence. This supports a model where early 5' splice recognition results from a network of interactions established by the splicing machinery with various regions of the pre-mRNA.
Figures



References
-
- Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. - PubMed
-
- Abovich N, Liao XC, Rosbash M. The yeast MUD2 protein: An interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 1994;8:843–854. - PubMed
-
- Berglund JA, Chua K, Abovich N, Reed R, Rosbash M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
