Nucleoid-independent identification of cell division sites in Escherichia coli
- PMID: 10074085
- PMCID: PMC93591
- DOI: 10.1128/JB.181.6.1900-1905.1999
Nucleoid-independent identification of cell division sites in Escherichia coli
Abstract
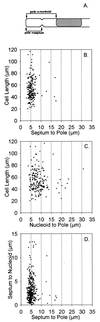
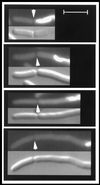
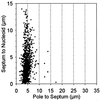
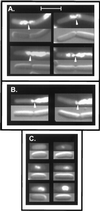
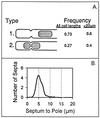
The mechanism used by Escherichia coli to determine the correct site for cell division is unknown. In this report, we have attempted to distinguish between a model in which septal position is determined by the position of the nucleoids and a model in which septal position is predetermined by a mechanism that does not involve nucleoid position. To do this, filaments with extended nucleoid-free regions adjacent to the cell poles were produced by simultaneous inactivation of cell division and DNA replication. The positions of septa that formed within the nucleoid-free zones after division was allowed to resume were then analyzed. The results showed that septa were formed at a uniform distance from cell poles when division was restored, with no relation to the distance from the nearest nucleoid. In some cells, septa were formed directly over nucleoids. These results are inconsistent with models that invoke nucleoid positioning as the mechanism for determining the site of division site formation.
Figures


References
-
- Åkerlund T, Bernander R, Nordström K. Cell division in Escherichia coli minB mutants. Mol Microbiol. 1992;6:2073–2083. - PubMed
-
- Cook W, Rothfield L. Early stages in development of the E. coli division site. Mol Microbiol. 1994;14:485–495. - PubMed
-
- Cook W, Rothfield L I. Development of potential division sites in FtsA− filaments of E. coli. Mol Microbiol. 1994;14:497–503. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

