The transmembrane domain of hepatitis C virus glycoprotein E1 is a signal for static retention in the endoplasmic reticulum
- PMID: 10074109
- PMCID: PMC104019
- DOI: 10.1128/JVI.73.4.2641-2649.1999
The transmembrane domain of hepatitis C virus glycoprotein E1 is a signal for static retention in the endoplasmic reticulum
Abstract
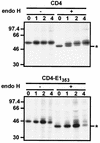
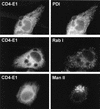
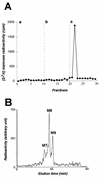
Hepatitis C virus (HCV) glycoproteins E1 and E2 assemble to form a noncovalent heterodimer which, in the cell, accumulates in the endoplasmic reticulum (ER). Contrary to what is observed for proteins with a KDEL or a KKXX ER-targeting signal, the ER localization of the HCV glycoprotein complex is due to a static retention in this compartment rather than to its retrieval from the cis-Golgi region. A static retention in the ER is also observed when E2 is expressed in the absence of E1 or for a chimeric protein containing the ectodomain of CD4 in fusion with the transmembrane domain (TMD) of E2. Although they do not exclude the presence of an intracellular localization signal in E1, these data do suggest that the TMD of E2 is an ER retention signal for HCV glycoprotein complex. In this study chimeric proteins containing the ectodomain of CD4 or CD8 fused to the C-terminal hydrophobic sequence of E1 were shown to be localized in the ER, indicating that the TMD of E1 is also a signal for ER localization. In addition, these chimeric proteins were not processed by Golgi enzymes, indicating that the TMD of E1 is responsible for true retention in the ER, without recycling through the Golgi apparatus. Together, these data suggest that at least two signals (TMDs of E1 and E2) are involved in ER retention of the HCV glycoprotein complex.
Figures





References
-
- Ahn K, Szczesna-Skorupa E, Kemper B. The amino-terminal 29 amino acids of cytochrome P450 2C1 are sufficient for retention in the endoplasmic reticulum. J Biol Chem. 1993;268:18726–18733. - PubMed
-
- Armstrong J, Patel S. The Golgi sorting domain of coronavirus E1 protein. J Cell Sci. 1991;98:567–575. - PubMed
-
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach M F, Ravazzola M, Amherdt M, Schekman R. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. - PubMed
-
- Bause E, Breuer W, Schweden J, Roeser R, Geyer R. Effect of substrate structure on the activity of Man9-mannosidase from pig liver involved in N-linked oligosaccharide processing. Eur J Biochem. 1992;208:451–457. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

