The ski7 antiviral protein is an EF1-alpha homolog that blocks expression of non-Poly(A) mRNA in Saccharomyces cerevisiae
- PMID: 10074137
- PMCID: PMC104047
- DOI: 10.1128/JVI.73.4.2893-2900.1999
The ski7 antiviral protein is an EF1-alpha homolog that blocks expression of non-Poly(A) mRNA in Saccharomyces cerevisiae
Abstract
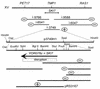
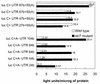
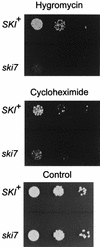
We mapped and cloned SKI7, a gene that negatively controls the copy number of L-A and M double-stranded RNA viruses in Saccharomyces cerevisiae. We found that it encodes a nonessential 747-residue protein with similarities to two translation factors, Hbs1p and EF1-alpha. The ski7 mutant was hypersensitive to hygromycin B, a result also suggesting a role in translation. The SKI7 product repressed the expression of nonpolyadenylated [non-poly(A)] mRNAs, whether capped or uncapped, thus explaining why Ski7p inhibits the propagation of the yeast viruses, whose mRNAs lack poly(A). The dependence of the Ski7p effect on 3' RNA structures motivated a study of the expression of capped non-poly(A) luciferase mRNAs containing 3' untranslated regions (3'UTRs) differing in length. In a wild-type strain, increasing the length of the 3'UTR increased luciferase expression due to both increased rates and duration of translation. Overexpression of Ski7p efficiently cured the satellite virus M2 due to a twofold-increased repression of non-poly(A) mRNA expression. Our experiments showed that Ski7p is part of the Ski2p-Ski3p-Ski8p antiviral system because a single ski7 mutation derepresses the expression of non-poly(A) mRNA as much as a quadruple ski2 ski3 ski7 ski8 mutation, and the effect of the overexpression of Ski7p is not obtained unless other SKI genes are functional. ski1/xrn1Delta ski2Delta and ski1/xrn1Delta ski7Delta mutants were viable but temperature sensitive for growth.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

