Genetic evidence that EBNA-1 is needed for efficient, stable latent infection by Epstein-Barr virus
- PMID: 10074147
- PMCID: PMC104057
- DOI: 10.1128/JVI.73.4.2974-2982.1999
Genetic evidence that EBNA-1 is needed for efficient, stable latent infection by Epstein-Barr virus
Abstract
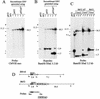
Replication and maintenance of the 170-kb circular chromosome of Epstein-Barr virus (EBV) during latent infection are generally believed to depend upon a single viral gene product, the nuclear protein EBNA-1. EBNA-1 binds to two clusters of sites at oriP, an 1, 800-bp sequence on the EBV genome which can support replication and maintenance of artificial plasmids introduced into cell lines that contain EBNA-1. To investigate the importance of EBNA-1 to latent infection by EBV, we introduced a frameshift mutation into the EBNA-1 gene of EBV by recombination along with a flanking selectable marker. EBV genomes carrying the frameshift mutation could be isolated readily after superinfecting EBV-positive cell lines, but not if recombinant virus was used to infect EBV-negative B-cell lines or to immortalize peripheral blood B cells. EBV mutants lacking almost all of internal repeat 3, which encode a repetitive glycine and alanine domain of EBNA-1, were generated in the same way and found to immortalize B cells normally. An EBNA-1-deficient mutant of EBV was isolated and found to be incapable of establishing a latent infection of the cell line BL30 at a detectable frequency, indicating that the mutant was less than 1% as efficient as an isogenic, EBNA-1-positive strain in this assay. The data indicate that EBNA-1 is required for efficient and stable latent infection by EBV under the conditions tested. Evidence from other studies now indicates that autonomous maintenance of the EBV chromosome during latent infection does not depend on the replication initiation function of oriP. It is therefore likely that the viral chromosome maintenance (segregation) function of oriP and EBNA-1 is what is required.
Figures


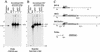


References
-
- Decker L L, Shankar P, Khan G, Freeman R B, Dezube B J, Lieberman J, Thorley-Lawson D A. The Kaposi sarcoma-associated herpesvirus (KSHV) is present as an intact latent genome in KS tissue but replicates in the peripheral blood mononuclear cells of KS patients. J Exp Med. 1996;184:283–288. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

