HMG protein family members stimulate human immunodeficiency virus type 1 and avian sarcoma virus concerted DNA integration in vitro
- PMID: 10074149
- PMCID: PMC104059
- DOI: 10.1128/JVI.73.4.2994-3003.1999
HMG protein family members stimulate human immunodeficiency virus type 1 and avian sarcoma virus concerted DNA integration in vitro
Abstract
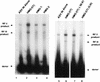
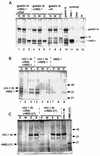
We have reconstituted concerted human immunodeficiency virus type 1 (HIV-1) integration in vitro with specially designed mini-donor HIV-1 DNA, a supercoiled plasmid acceptor, purified bacterium-derived HIV-1 integrase (IN), and host HMG protein family members. This system is comparable to one previously described for avian sarcoma virus (ASV) (A. Aiyar et al., J. Virol. 70:3571-3580, 1996) that was stimulated by the presence of HMG-1. Sequence analyses of individual HIV-1 integrants showed loss of 2 bp from the ends of the donor DNA and almost exclusive 5-bp duplications of the acceptor DNA at the site of integration. All of the integrants sequenced were inserted into different sites in the acceptor. These are the features associated with integration of viral DNA in vivo. We have used the ASV and HIV-1 reconstituted systems to compare the mechanism of concerted DNA integration and examine the role of different HMG proteins in the reaction. Of the three HMG proteins examined, HMG-1, HMG-2, and HMG-I(Y), the products formed in the presence of HMG-I(Y) for both systems most closely match those observed in vivo. Further analysis of HMG-I(Y) mutants demonstrates that the stimulation of integration requires an HMG-I(Y) domain involved in DNA binding. While complexes containing HMG-I(Y), ASV IN, and donor DNA can be detected in gel shift experiments, coprecipitation experiments failed to demonstrate stable interactions between HMG-I(Y) and ASV IN or between HMG-I(Y) and HIV-1 IN.
Figures


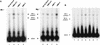


References
-
- Andrake M D, Skalka A M. Multimerization determinants reside in both the catalytic core and C terminus of avian sarcoma virus integrase. J Biol Chem. 1995;270:29299–29306. - PubMed
-
- Asante-Appiah E, Skalka A M. A metal-induced conformational change and activation of HIV-1 integrase. J Biol Chem. 1997;272:16196–16205. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. 2nd ed. New York, N.Y: Greene Publishing Associates and John Wiley & Sons; 1992.
-
- Bustin M, Lehn D A, Landsman D. Structural features of the HMG chromosomal proteins and their genes. Biochim Biophys Acta. 1990;1049:231–243. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

