ERV-L elements: a family of endogenous retrovirus-like elements active throughout the evolution of mammals
- PMID: 10074184
- PMCID: PMC104094
- DOI: 10.1128/JVI.73.4.3301-3308.1999
ERV-L elements: a family of endogenous retrovirus-like elements active throughout the evolution of mammals
Abstract
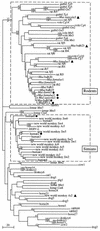
We have previously identified in the human genome a family of 200 endogenous retrovirus-like elements, the HERV-L elements, disclosing similarities with the foamy retroviruses and which might be the evolutionary intermediate between classical intracellular retrotransposons and infectious retroviruses. Southern blot analysis of a large series of mammalian genomic DNAs shows that HERV-L-related elements-so-called ERV-L-are present among all placental mammals, suggesting that ERV-L elements were already present at least 70 million years ago. Most species exhibit a low copy number of ERV-L elements (from 10 to 30), while simians (not prosimians) and mice (not rats) have been subjected to bursts resulting in increases in the number of copies up to 200. The burst of copy number in primates can be dated to shortly after the prosimian and simian branchpoint, 45 to 65 million years ago, whereas murine species have been subjected to two much more recent bursts (less than 10 million years ago), occurring after the Mus/Rattus split. We have amplified and sequenced 360-bp ERV-L internal fragments of the highly conserved pol gene from a series of 22 mammalian species. These sequences exhibit high percentages of identity (57 to 99%) with the murine fully coding MuERV-L element. Phylogenetic analyses allowed the establishment of a plausible evolutionary scheme for ERV-L elements, which accounts for the high level of sequence conservation and the widespread dispersion among mammals.
Figures



References
-
- Adachi J, Hasegawa M. Computer monographs. Vol. 28. Tokyo, Japan: Institute of Statistical Mathematics; 1996. MOLPHY: programs for molecular phylogenetics based on maximum likelihood, version 2.3.
-
- Bénit L, de Parseval N, Casella J-F, Callebaut I, Cordonnier A, Heidmann T. Cloning of a new murine endogenous retrovirus, MuERV-L, with strong similarity to the human HERV-L element and with a gag coding sequence closely related to the Fv1 restriction gene. J Virol. 1997;71:5652–5257. - PMC - PubMed
-
- Best S, le Tissier P, Towers G, Stoye J P. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. - PubMed
-
- Boursot P, Auffray J C, Britton-Davidian J, Bonhomme F. The evolution of house mice. Annu Rev Ecol Syst. 1993;24:119–152.
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

