Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras
- PMID: 10074190
- PMCID: PMC104100
- DOI: 10.1128/JVI.73.4.3359-3365.1999
Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras
Abstract
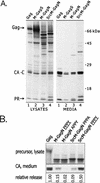
Little is known about the mechanisms used by enveloped viruses to separate themselves from the cell surface at the final step of budding. However, small sequences in the Gag proteins of several retroviruses (L domains) have been implicated in this process. A sequence has been identified in the M proteins of rhabdoviruses that closely resembles the PPPPY motif in the L domain of Rous sarcoma virus (RSV), an avian retrovirus. To evaluate whether the PPPY sequence in vesicular stomatitis virus (VSV) M protein has an activity analogous to that of the retroviral sequence, M-Gag chimeras were characterized. The N-terminal 74 amino acids of the VSV (Indiana) M protein, including the PPPY motif, was able to replace the L domain of RSV Gag and allow the assembly and release of virus-like particles. Alanine substitutions in the VSV PPPY motif severely compromised the budding activity of this hybrid protein but not that of another chimera which also contained the RSV PPPPY sequence. We conclude that this VSV sequence is functionally homologous to the RSV L domain in promoting virus particle release, making this the first example of such an activity in a virus other than a retrovirus. Both the RSV and VSV motifs have been shown to interact in vitro with certain cellular proteins that contain a WW interaction module, suggesting that the L domains are sites of interaction with unknown host machinery involved in virus release.
Figures



References
-
- Chen H I, Einbond A, Kwak S-J, Linn H, Koepf E, Peterson S, Kelly J W, Sudol M. Characterization of the WW domain of human Yes-associated protein and its polyproline-containing ligands. J Biol Chem. 1997;272:17070–17077. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

