Clinical significance of expression of human cytomegalovirus pp67 late transcript in heart, lung, and bone marrow transplant recipients as determined by nucleic acid sequence-based amplification
- PMID: 10074499
- PMCID: PMC88622
- DOI: 10.1128/JCM.37.4.902-911.1999
Clinical significance of expression of human cytomegalovirus pp67 late transcript in heart, lung, and bone marrow transplant recipients as determined by nucleic acid sequence-based amplification
Abstract
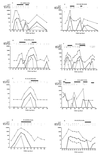
Human cytomegalovirus (HCMV) infection was monitored retrospectively by qualitative determination of pp67 mRNA (a late viral transcript) by nucleic acid sequence-based amplification (NASBA) in a series of 50 transplant recipients, including 26 solid-organ (11 heart and 15 lung) transplant recipients (SOTRs) and 24 bone marrow transplant recipients (BMTRs). NASBA results were compared with those obtained by prospective quantitation of HCMV viremia and antigenemia and retrospective quantitation of DNA in leukocytes (leukoDNAemia). On the whole, 29 patients were NASBA positive, whereas 10 were NASBA negative, and the blood of 11 patients remained HCMV negative. NASBA detected HCMV infection before quantitation of viremia did but after quantitation of leukoDNAemia and antigenemia did. In NASBA-positive blood samples, median levels of viremia, antigenemia, and leukoDNAemia were significantly higher than the relevant levels detected in NASBA-negative HCMV-positive blood samples. By using the quantitation of leukoDNAemia as the "gold standard," the analytical sensitivity (47.3%), as well as the negative predictive value (68. 3%), of NASBA for the diagnosis of HCMV infection intermediate between that of antigenemia quantitation (analytical sensitivity, 72. 3%) and that of viremia quantitation (analytical sensitivity, 28.7%), while the specificity and the positive predictive value were high (90 to 100%). However, with respect to the clinically relevant antigenemia cutoff of >/=100 used in this study for the initiation of preemptive therapy in SOTRs with reactivated HCMV infection, the clinical sensitivity of NASBA reached 100%, with a specificity of 68. 9%. Upon the initiation of antigenemia quantitation-guided treatment, the actual median antigenemia level was 158 (range, 124 to 580) in SOTRs who had reactivated infection and who presented with NASBA positivity 3.5 +/- 2.6 days in advance and 13.5 (range, 1 to 270) in the group that included BMTRs and SOTRs who had primary infection (in whom treatment was initiated upon the first confirmation of detection of HCMV in blood) and who presented with NASBA positivity 2.0 +/- 5.1 days later. Following antiviral treatment, the durations of the presence of antigenemia and pp67 mRNA in blood were found to be similar. In conclusion, monitoring of the expression of HCMV pp67 mRNA appears to be a promising, well-standardized tool for determination of the need for the initiation and termination of preemptive therapy. Its overall clinical impact should be analyzed in future prospective studies.
Figures
References
-
- Bitsch A, Kirchner H, Dupke R, Bein G. Cytomegalovirus transcripts in peripheral blood leukocytes of actively infected transplant patients detected by reverse-transcription-polymerase chain reaction. J Infect Dis. 1993;167:740–743. - PubMed
-
- Blok M J, Goossens V J, Vanherle S J V, Top B, Tacken N, Middeldorp J M, Christiaans M H L, van Hooff J P, Bruggeman C A. Diagnostic value of monitoring human cytomegalovirus late pp67 mRNA expression in renal-allograft recipients by nucleic acid sequence-based amplification. J Clin Microbiol. 1998;36:1341–1346. - PMC - PubMed
-
- Boivin G, Olson C A, Quirk M R, St.-Cyr S M, Jordan M C. Quantitation of cytomegalovirus glycoprotein H gene in cells using competitive PCR and a rapid fluorescent-based detection system. J Virol Methods. 1995;51:329–342. - PubMed
-
- Compton J. Nucleic acid sequence-based amplification. Nature. 1991;350:91–92. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

