Species identification and strain differentiation of dermatophyte fungi by analysis of ribosomal-DNA intergenic spacer regions
- PMID: 10074504
- PMCID: PMC88627
- DOI: 10.1128/JCM.37.4.931-936.1999
Species identification and strain differentiation of dermatophyte fungi by analysis of ribosomal-DNA intergenic spacer regions
Abstract
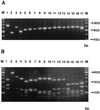
Restriction fragment length polymorphisms (RFLPs) identified in the ribosomal-DNA (rDNA) repeat were used for molecular strain differentiation of the dermatophyte fungus Trichophyton rubrum. The polymorphisms were detected by hybridization of EcoRI-digested T. rubrum genomic DNAs with a probe amplified from the small-subunit (18S) rDNA and adjacent internal transcribed spacer (ITS) regions. The rDNA RFLPs mapped to the nontranscribed spacer (NTS) region of the rDNA repeat and appeared similar to those caused by short repetitive sequences in the intergenic spacers of other fungi. Fourteen individual RFLP patterns (DNA types A to N) were recognized among 50 random clinical isolates of T. rubrum. A majority of strains (19 of 50 [38%]) were characterized by one RFLP pattern (DNA type A), and four types (DNA types A to D) accounted for 78% (39 of 50) of all strains. The remaining types (DNA types E to N) were represented by one or two isolates only. A rapid and simple method was also developed for molecular species identification of dermatophyte fungi. The contiguous ITS and 5.8S rDNA regions were amplified from 17 common dermatophyte species by using the universal primers ITS 1 and ITS 4. Digestion of the amplified ITS products with the restriction endonuclease MvaI produced unique and easily identifiable fragment patterns for a majority of species. However, some closely related taxon pairs, such as T. rubrum-T. soudanense and T. quinkeanum-T. schoenlenii could not be distinguished. We conclude that RFLP analysis of the NTS and ITS intergenic regions of the rDNA repeat is a valuable technique both for molecular strain differentiation of T. rubrum and for species identification of common dermatophyte fungi.
Figures
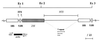

Similar articles
-
Molecular characterization of dermatophytes isolated from cattle in Plateau State, Nigeria.Vet Microbiol. 2018 Jun;219:212-218. doi: 10.1016/j.vetmic.2018.04.022. Epub 2018 Apr 20. Vet Microbiol. 2018. PMID: 29778198
-
Restriction fragment length polymorphism analysis of ribosomal DNA intergenic regions is useful for differentiating strains of Trichophyton mentagrophytes.J Clin Microbiol. 2003 Oct;41(10):4583-8. doi: 10.1128/JCM.41.10.4583-4588.2003. J Clin Microbiol. 2003. PMID: 14532186 Free PMC article.
-
Direct species identification of common pathogenic dermatophyte fungi in clinical specimens by semi-nested PCR and restriction fragment length polymorphism.Mycopathologia. 2008 Oct;166(4):203-8. doi: 10.1007/s11046-008-9130-3. Epub 2008 Jun 4. Mycopathologia. 2008. PMID: 18523862
-
Molecular identification and strain typing of dermatophyte fungi.Nihon Ishinkin Gakkai Zasshi. 2001;42(1):7-10. doi: 10.3314/jjmm.42.7. Nihon Ishinkin Gakkai Zasshi. 2001. PMID: 11173329 Review.
-
Strain differentiation of dermatophytes.Mycopathologia. 2008 Nov-Dec;166(5-6):319-33. doi: 10.1007/s11046-008-9108-1. Epub 2008 May 14. Mycopathologia. 2008. PMID: 18478358 Review.
Cited by
-
Transcriptional profiles of the response to ketoconazole and amphotericin B in Trichophyton rubrum.Antimicrob Agents Chemother. 2007 Jan;51(1):144-53. doi: 10.1128/AAC.00755-06. Epub 2006 Oct 23. Antimicrob Agents Chemother. 2007. PMID: 17060531 Free PMC article.
-
Variations in ribosomal DNA copy numbers in a genome of Trichophyton interdigitale.Mycoses. 2020 Oct;63(10):1133-1140. doi: 10.1111/myc.13163. Epub 2020 Sep 2. Mycoses. 2020. PMID: 32783279 Free PMC article.
-
Variation in restriction fragment length polymorphisms among serial isolates from patients with Trichophyton rubrum infection.J Clin Microbiol. 2001 Sep;39(9):3260-6. doi: 10.1128/JCM.39.9.3260-3266.2001. J Clin Microbiol. 2001. PMID: 11526160 Free PMC article.
-
Multiplex PCR using internal transcribed spacer 1 and 2 regions for rapid detection and identification of yeast strains.J Clin Microbiol. 2001 Oct;39(10):3617-22. doi: 10.1128/JCM.39.10.3617-3622.2001. J Clin Microbiol. 2001. PMID: 11574582 Free PMC article.
-
Application of polymerase chain reaction (PCR) and PCR based restriction fragment length polymorphism for detection and identification of dermatophytes from dermatological specimens.Indian J Dermatol. 2008 Jan;53(1):15-20. doi: 10.4103/0019-5154.39735. Indian J Dermatol. 2008. PMID: 19967012 Free PMC article.
References
-
- Berbee M L, Yoshimura A, Sugiyama J, Taylor J W. Is Penicillium monophyletic—an evaluation of phylogeny in the family Trichocomaceae from 18S, 5.8S and ITS ribosomal DNA sequence data. Mycologia. 1995;87:210–222.
-
- Bowman B H, Taylor J W, White T J. Molecular evolution of the fungi: human pathogens. Mol Biol Evol. 1992;9:893–904. - PubMed
-
- Carlotti A, Srikantha T, Schröppel K, Kvaal C, Villard J, Soll D R. A novel repeat sequence (CKRS-1) containing a tandemly repeated sub-element (kre) accounts for differences between Candida krusei strains fingerprinted with the probe CkF1,2. Curr Genet. 1997;31:255–263. - PubMed
-
- Evans E G V. Causative pathogens in onychomycosis and the possibility of treatment resistance: a review. J Am Acad Dermatol. 1998;38:S32–S56. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

