Modulation of CREB binding protein function by the promyelocytic (PML) oncoprotein suggests a role for nuclear bodies in hormone signaling
- PMID: 10077561
- PMCID: PMC15819
- DOI: 10.1073/pnas.96.6.2627
Modulation of CREB binding protein function by the promyelocytic (PML) oncoprotein suggests a role for nuclear bodies in hormone signaling
Abstract
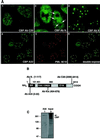
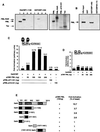
Disaggregation of the spherical nuclear bodies termed promyelocytic (PML) oncogenic domains (PODs) is a characteristic of acute promyelocytic leukemia. Here, we demonstrate that the cAMP enhancer binding protein (CREB)-binding protein (CBP) associates with PML in vitro and is recruited to the PODs in vivo. Through its association with CBP, wild-type PML dramatically stimulates nuclear receptor transcriptional activity. These results demonstrate that a fraction of CBP is compartmentalized to the POD through its association with PML and thus suggest that PML and other POD-associated proteins may play an unexpectedly broad role in aspects of transcriptional regulation and human disease.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

