Transcriptional activation by artificial recruitment in yeast is influenced by promoter architecture and downstream sequences
- PMID: 10077568
- PMCID: PMC15826
- DOI: 10.1073/pnas.96.6.2668
Transcriptional activation by artificial recruitment in yeast is influenced by promoter architecture and downstream sequences
Abstract
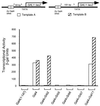
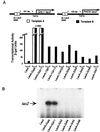
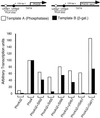
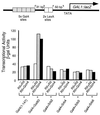
The idea that recruitment of the transcriptional machinery to a promoter suffices for gene activation is based partly on the results of "artificial recruitment" experiments performed in vivo. Artificial recruitment can be effected by a "nonclassical" activator comprising a DNA-binding domain fused to a component of the transcriptional machinery. Here we show that activation by artificial recruitment in yeast can be sensitive to any of three factors: position of the activator-binding elements, sequence of the promoter, and coding sequences downstream of the promoter. In contrast, classical activators worked efficiently at all promoters tested. In all cases the "artificial recruitment" fusions synergized well with classical activators. A classical activator evidently differs from a nonclassical activator in that the former can touch multiple sites on the transcriptional machinery, and we propose that that difference accounts for the broader spectrum of activity of the typical classical activator. A similar conclusion is reached from studies in mammalian cells in the accompanying paper [Nevado, J., Gaudreau, L., Adam, M. & Ptashne, M. (1999) Proc. Natl. Acad. Sci. USA 96, 2674-2677].
Figures


Similar articles
-
Transcriptional activation by artificial recruitment in mammalian cells.Proc Natl Acad Sci U S A. 1999 Mar 16;96(6):2674-7. doi: 10.1073/pnas.96.6.2674. Proc Natl Acad Sci U S A. 1999. PMID: 10077569 Free PMC article.
-
Recruitment of TBP or TFIIB to a promoter proximal position leads to stimulation of RNA polymerase II transcription without activator proteins both in vivo and in vitro.Biochem Biophys Res Commun. 1999 Mar 5;256(1):45-51. doi: 10.1006/bbrc.1999.0280. Biochem Biophys Res Commun. 1999. PMID: 10066420
-
Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme.Nature. 1999 Jun 10;399(6736):609-13. doi: 10.1038/21239. Nature. 1999. PMID: 10376605
-
Activation of the Gal1 gene of yeast by pairs of 'non-classical' activators.Curr Biol. 2004 Sep 21;14(18):1675-9. doi: 10.1016/j.cub.2004.09.025. Curr Biol. 2004. PMID: 15380071
-
Yeast transcriptional regulatory mechanisms.Annu Rev Genet. 1995;29:651-74. doi: 10.1146/annurev.ge.29.120195.003251. Annu Rev Genet. 1995. PMID: 8825489 Review.
Cited by
-
A target essential for the activity of a nonacidic yeast transcriptional activator.Proc Natl Acad Sci U S A. 2002 Jun 25;99(13):8591-6. doi: 10.1073/pnas.092263499. Proc Natl Acad Sci U S A. 2002. PMID: 12084920 Free PMC article.
-
Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators.Genetics. 2011 Nov;189(3):705-36. doi: 10.1534/genetics.111.127019. Genetics. 2011. PMID: 22084422 Free PMC article. Review.
-
An artificial transcriptional activating region with unusual properties.Proc Natl Acad Sci U S A. 2000 Feb 29;97(5):1988-92. doi: 10.1073/pnas.040573197. Proc Natl Acad Sci U S A. 2000. PMID: 10681438 Free PMC article.
-
Artificial recruitment of TFIID, but not RNA polymerase II holoenzyme, activates transcription in mammalian cells.Mol Cell Biol. 2000 Jun;20(12):4350-8. doi: 10.1128/MCB.20.12.4350-4358.2000. Mol Cell Biol. 2000. PMID: 10825198 Free PMC article.
-
Revisiting the model for coactivator recruitment: Med15 can select its target sites independent of promoter-bound transcription factors.Nucleic Acids Res. 2024 Nov 11;52(20):12093-12111. doi: 10.1093/nar/gkae718. Nucleic Acids Res. 2024. PMID: 39187372 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

