The schizosaccharomyces pombe dim1(+) gene interacts with the anaphase-promoting complex or cyclosome (APC/C) component lid1(+) and is required for APC/C function
- PMID: 10082519
- PMCID: PMC84046
- DOI: 10.1128/MCB.19.4.2535
The schizosaccharomyces pombe dim1(+) gene interacts with the anaphase-promoting complex or cyclosome (APC/C) component lid1(+) and is required for APC/C function
Abstract
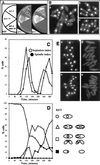
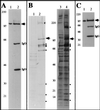
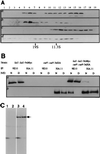
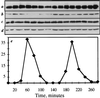
The Schizosaccharomyces pombe dim1(+) gene is required for entry into mitosis and for chromosome segregation during mitosis. To further understand dim1p function, we undertook a synthetic lethal screen with the temperature-sensitive dim1-35 mutant and isolated lid (for lethal in dim1-35) mutants. Here, we describe the temperature-sensitive lid1-6 mutant. At the restrictive temperature of 36 degrees C, lid1-6 mutant cells arrest with a "cut" phenotype similar to that of cut4 and cut9 mutants. An epitope-tagged version of lid1p is a component of a multiprotein approximately 20S complex; the presence of lid1p in this complex depends upon functional cut9(+). lid1p-myc coimmunoprecipitates with several other proteins, including cut9p and nuc2p, and the presence of cut9p in a 20S complex depends upon the activity of lid1(+). Further, lid1(+) function is required for the multiubiquitination of cut2p, an anaphase-promoting complex or cyclosome (APC/C) target. Thus, lid1p is a component of the S. pombe APC/C. In dim1 mutants, the abundances of lid1p and the APC/C complex decline significantly, and the ubiquitination of an APC/C target is abolished. These data suggest that at least one role of dim1p is to maintain or establish the steady-state level of the APC/C.
Figures






References
-
- Bahler J, Wu J Q, Longtine M S, Shah N G, McKenzie III A, Steever A B, Wach A, Philippsen P, Pringle J R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. - PubMed
-
- Balasubramanian M K, McCollum D, Gould K L. Cytokinesis in fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1997;283:494–506. - PubMed
-
- Barbet N, Muriel W J, Carr A M. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. Gene. 1992;114:59–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
