Rpb7 can interact with RNA polymerase II and support transcription during some stresses independently of Rpb4
- PMID: 10082533
- PMCID: PMC84060
- DOI: 10.1128/MCB.19.4.2672
Rpb7 can interact with RNA polymerase II and support transcription during some stresses independently of Rpb4
Abstract
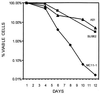
Rpb4 and Rpb7 are two yeast RNA polymerase II (Pol II) subunits whose mechanistic roles have recently started to be deciphered. Although previous data suggest that Rpb7 can stably interact with Pol II only as a heterodimer with Rpb4, RPB7 is essential for viability, whereas RPB4 is essential only during some stress conditions. To resolve this discrepancy and to gain a better understanding of the mode of action of Rpb4, we took advantage of the inability of cells lacking RPB4 (rpb4Delta, containing Pol IIDelta4) to grow above 30 degrees C and screened for genes whose overexpression could suppress this defect. We thus discovered that overexpression of RPB7 could suppress the inability of rpb4Delta cells to grow at 34 degrees C (a relatively mild temperature stress) but not at higher temperatures. Overexpression of RPB7 could also partially suppress the cold sensitivity of rpb4Delta strains and fully suppress their inability to survive a long starvation period (stationary phase). Notably, however, overexpression of RPB4 could not override the requirement for RPB7. Consistent with the growth phenotype, overexpression of RPB7 could suppress the transcriptional defect characteristic of rpb4Delta cells during the mild, but not during a more severe, heat shock. We also demonstrated, through two reciprocal coimmunoprecipitation experiments, a stable interaction of the overproduced Rpb7 with Pol IIDelta4. Nevertheless, fewer Rpb7 molecules interacted with Pol IIDelta4 than with wild-type Pol II. Thus, a major role of Rpb4 is to augment the interaction of Rpb7 with Pol II. We suggest that Pol IIDelta4 contains a small amount of Rpb7 that is sufficient to support transcription only under nonstress conditions. When RPB7 is overexpressed, more Rpb7 assembles with Pol IIDelta4, enough to permit appropriate transcription also under some stress conditions.
Figures





References
-
- Acker J, de Graaff M, Cheynel I, Khazak V, Kedinger C, Vigneron M. Interactions between the human RNA polymerase II subunits. J Biol Chem. 1997;272:16815–16821. - PubMed
-
- Asturias F J, Meredith G D, Poglitsch C L, Kornberg R D. Two conformations of RNA polymerase II revealed by electron crystallography. J Mol Biol. 1997;272:536–540. - PubMed
-
- Becker D M, Guarente L. High-efficiency transformation of yeast by electroporation. Methods Enzymol. 1991;194:182–187. - PubMed
-
- Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. - PubMed
-
- Choder M. A general topoisomerase I-dependent transcriptional repression in the stationary phase in yeast. Genes Dev. 1991;5:2315–2326. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
