Reciprocal interaction between two cellular proteins, Puralpha and YB-1, modulates transcriptional activity of JCVCY in glial cells
- PMID: 10082537
- PMCID: PMC84064
- DOI: 10.1128/MCB.19.4.2712
Reciprocal interaction between two cellular proteins, Puralpha and YB-1, modulates transcriptional activity of JCVCY in glial cells
Abstract
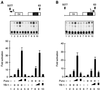
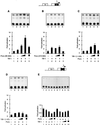
Cross communication between regulatory proteins is an important event in the control of eukaryotic gene transcription. Here we have examined the structural and functional interaction between two cellular regulatory proteins, YB-1 and Puralpha, on the 23-bp sequence element derived from the enhancer-promoter of the human polyomavirus JCV. YB-1 and Puralpha are single-stranded DNA binding proteins which recognize C/T- and GC/GA-rich sequences, respectively. Results from band shift studies demonstrated that while both proteins interact directly with their DNA target sequences within the 23-bp motif, each protein can regulate the association of the other one with the DNA. Affinity chromatography and coimmunoprecipitation provide evidence for a direct interaction between Puralpha and YB-1 in the absence of the DNA sequence. Ectopic expression of YB-1 and Puralpha in glial cells synergistically stimulated viral promoter activity via the 23-bp sequence element. Results from mutational studies revealed that residues between amino acids 75 and 203 of YB-1 and between amino acids 85 and 215 of Puralpha are important for the interaction between these two proteins. Functional studies with glial cells indicated that the region within Puralpha which mediates its association with YB-1 and binding to the 23-bp sequence is important for the observed activation of the JCV promoter by the Puralpha and YB-1 proteins. The results of this study suggest that the cooperative interaction between YB-1 and Puralpha mediates the synergistic activation of the human polyomavirus JCV genome by these cellular proteins. The importance of these findings for cellular and viral genes which are regulated by Puralpha and YB-1 is discussed.
Figures
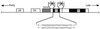




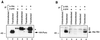


References
-
- Ahmed S, Rappaport J, Tada H, Kerr D, Khalili K. A nuclear protein derived from brain cells stimulates transcription of the human neurotropic virus promoter, JCVE, in vitro. J Biol Chem. 1990;265:13899–13905. - PubMed
-
- Asakuno K, Kohno K, Uchiumi T, Kubo T, Sato S, Isono M, Kuwano M. Involvement of a DNA binding protein, MDR-NF1/YB-1, in human MDR1 gene expression by actinomycin D. Biochem Biophys Res Commun. 1994;199:1428–1435. - PubMed
-
- Ausubel F, Brent R, Kingston R E, Moar D D, Siedman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1989.
-
- Bargou R C, Jurchott K, Wagener C, Bergmann S, Metzner S, Bommert K, Mapara M Y, Winzer K J, Dietel M, Dorken B, Royer H D. Nuclear localization and increased levels of transcription factor YB-1 in primary human breast cancers are associated with intrinsic MDR1 gene expression. Nat Med. 1997;3:447–450. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
