Binding of Gal4p and bicoid to nucleosomal sites in yeast in the absence of replication
- PMID: 10082565
- PMCID: PMC84092
- DOI: 10.1128/MCB.19.4.2977
Binding of Gal4p and bicoid to nucleosomal sites in yeast in the absence of replication
Abstract
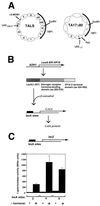
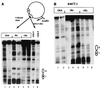
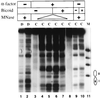
The yeast transcriptional activator Gal4p can bind to sites in nucleosomal DNA in vivo which it is unable to access in vitro. One event which could allow proteins to bind to otherwise inaccessible sites in chromatin in living cells is DNA replication. To determine whether replication is required for Gal4p to bind to nucleosomal sites in yeast, we have used previously characterized chromatin reporters in which Gal4p binding sites are incorporated into nucleosomes. We find that Gal4p is able to perturb nucleosome positioning via nucleosomal binding sites in yeast arrested either in G1, with alpha-factor, or in G2/M, with nocodazole. Similar results were obtained whether Gal4p synthesis was induced from the endogenous promoter by growth in galactose medium or by an artificial, hormone-inducible system. We also examined binding of the Drosophila transcriptional activator Bicoid, which belongs to the homeodomain class of transcription factors. We show that Bicoid, like Gal4p, can bind to nucleosomal sites in SWI+ and swi1Delta yeast and in the absence of replication. Our results indicate that some feature of the intracellular environment other than DNA replication or the SWI-SNF complex permits factor access to nucleosomal sites.
Figures
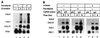



References
-
- Aparicio O, Gottschling D M. Overcoming telomeric silencing: a trans-activator competes to establish gene expression in a cell cycle-dependent way. Genes Dev. 1994;8:1133–1146. - PubMed
-
- Axelrod J D, Reagan M S, Majors J. GAL4 disrupts a repressing nucleosome during activation of GAL1 transcription in vivo. Genes Dev. 1993;7:857–869. - PubMed
-
- Balasubramanian, B., and R. H. Morse. Unpublished results.
-
- Barton M C, Emerson B M. Regulated expression of the β-globin gene locus in synthetic nuclei. Genes Dev. 1994;8:2453–2465. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
