Xist yeast artificial chromosome transgenes function as X-inactivation centers only in multicopy arrays and not as single copies
- PMID: 10082582
- PMCID: PMC84109
- DOI: 10.1128/MCB.19.4.3156
Xist yeast artificial chromosome transgenes function as X-inactivation centers only in multicopy arrays and not as single copies
Abstract
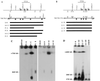
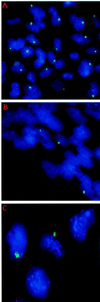
X-chromosome inactivation in female mammals is controlled by the X-inactivation center (Xic). This locus is required for inactivation in cis and is thought to be involved in the counting process which ensures that only a single X chromosome remains active per diploid cell. The Xist gene maps to the Xic region and has been shown to be essential for inactivation in cis. Transgenesis represents a stringent test for defining the minimal region that can carry out the functions attributed to the Xic. Although YAC and cosmid Xist-containing transgenes have previously been reported to be capable of cis inactivation and counting, the transgenes were all present as multicopy arrays and it was unclear to what extent individual copies are functional. Using two different yeast artificial chromosomes (YACs), we have found that single-copy transgenes, unlike multicopy arrays, can induce neither inactivation in cis nor counting. These results demonstrate that despite their large size and the presence of Xist, the YACs that we have tested lack sequences critical for autonomous function with respect to X inactivation.
Figures



References
-
- Ainscough J F-X, Koide T, Tada M, Barton S, Surani A. Imprinting of Igf2 and H19 from a 130 kb YAC transgene. Development. 1997;124:3621–3632. - PubMed
-
- Andrulis E D, Neiman A M, Zappulla D C, Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. - PubMed
-
- Arnaud, D. Unpublished data.
-
- Bartolomei M S, Webber A L, Brunkow M E, Tilghman S M. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Mol Cell Biol. 1993;7:1663–1673. - PubMed
-
- Borsani B, Tonlorenzi R, Simmler M-C, Dandolo L, Arnaud D, Capra V, Grompe M, Pizzuti A, Muzni D, Lawrence C, Willard H F, Avner P, Ballabio A. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991;351:325–329. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
