5-azacytidine induces transgene silencing by DNA methylation in Chinese hamster cells
- PMID: 10082586
- PMCID: PMC84113
- DOI: 10.1128/MCB.19.4.3198
5-azacytidine induces transgene silencing by DNA methylation in Chinese hamster cells
Abstract
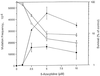
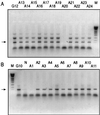
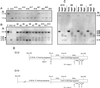
The cytosine analog 5-azacytidine (5-AzaC) is a demethylating agent that is also known to induce mutagenesis in mammalian cells. In this study, the mutagenic potential of this drug was tested in the G10 and G12 transgenic Chinese hamster cell lines, which have a single bacterial gpt gene integrated into the genome at different sites, with its expression driven by a simian virus 40 (SV40) promoter. We show that the mutation frequencies following a 48-h exposure to different concentrations of 5-AzaC were 10 to 20 times higher than those of any of the other numerous mutagens that have been tested in the G10-G12 system. Moreover, the mutation frequencies were much higher in the G10 cell line than in the G12 cells. Detailed molecular analysis of the 6-thioguanine (6-TG)-resistant variants demonstrated that transgene silencing by de novo DNA methylation and increased chromatin condensation in the SV40 promoter was the major factor responsible for this high level of 6-TG resistance. As would be expected, exposure to 5-AzaC lowered the overall genomic DNA methylation levels, but it unexpectedly caused hypermethylation and increased chromatin condensation of the transgene in both the G10 and G12 cell lines. These results provide the first evidence that 5-AzaC may also induce transgene-specific DNA methylation, a phenomenon that can further be used for the elucidation of the mechanism that controls silencing of foreign DNA.
Figures



References
-
- Amacher D, Turner G. The mutagenicity of 5-azacytidine and other inhibitors of replicative DNA synthesis in the L5178Y mouse lymphoma cell. Mutat Res. 1987;176:123–131. - PubMed
-
- Bender C M, Pao M M, Jones P A. Inhibition of DNA methylation by 5-aza-2′-deoxycytidine suppresses the growth of human tumor cell lines. Cancer Res. 1998;58:95–101. - PubMed
-
- Call K, Jensen J, Liber H, Thilly W. Studies of mutagenicity and clastogenicity of 5-azacytidine in human lymphoblasts and Salmonella typhimurium. Mutat Res. 1986;160:249–257. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
