Control of mitotic spindle position by the Saccharomyces cerevisiae formin Bni1p
- PMID: 10085293
- PMCID: PMC2148193
- DOI: 10.1083/jcb.144.5.947
Control of mitotic spindle position by the Saccharomyces cerevisiae formin Bni1p
Abstract
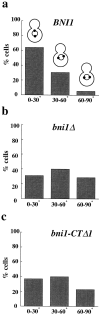
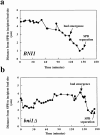
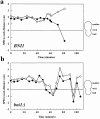
Alignment of the mitotic spindle with the axis of cell division is an essential process in Saccharomyces cerevisiae that is mediated by interactions between cytoplasmic microtubules and the cell cortex. We found that a cortical protein, the yeast formin Bni1p, was required for spindle orientation. Two striking abnormalities were observed in bni1Delta cells. First, the initial movement of the spindle pole body (SPB) toward the emerging bud was defective. This phenotype is similar to that previously observed in cells lacking the kinesin Kip3p and, in fact, BNI1 and KIP3 were found to be in the same genetic pathway. Second, abnormal pulling interactions between microtubules and the cortex appeared to cause preanaphase spindles in bni1Delta cells to transit back and forth between the mother and the bud. We therefore propose that Bni1p may localize or alter the function of cortical microtubule-binding sites in the bud. Additionally, we present evidence that other bipolar bud site determinants together with cortical actin are also required for spindle orientation.
Figures





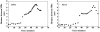



Similar articles
-
Dynamic positioning of mitotic spindles in yeast: role of microtubule motors and cortical determinants.Mol Biol Cell. 2000 Nov;11(11):3949-61. doi: 10.1091/mbc.11.11.3949. Mol Biol Cell. 2000. PMID: 11071919 Free PMC article.
-
Kinesin-related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration.J Cell Biol. 1997 Sep 8;138(5):1023-40. doi: 10.1083/jcb.138.5.1023. J Cell Biol. 1997. PMID: 9281581 Free PMC article.
-
Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins.J Cell Biol. 1997 Sep 8;138(5):1041-53. doi: 10.1083/jcb.138.5.1041. J Cell Biol. 1997. PMID: 9281582 Free PMC article.
-
Spindle polarity in S. cerevisiae: MEN can tell.Cell Cycle. 2002 Sep-Oct;1(5):308-11. doi: 10.4161/cc.1.5.143. Cell Cycle. 2002. PMID: 12461289 Review.
-
Control of spindle polarity and orientation in Saccharomyces cerevisiae.Trends Cell Biol. 2001 Apr;11(4):160-6. doi: 10.1016/s0962-8924(01)01954-7. Trends Cell Biol. 2001. PMID: 11306295 Review.
Cited by
-
Yeast formin Bni1p has multiple localization regions that function in polarized growth and spindle orientation.Mol Biol Cell. 2012 Feb;23(3):412-22. doi: 10.1091/mbc.E11-07-0631. Epub 2011 Dec 7. Mol Biol Cell. 2012. PMID: 22160598 Free PMC article.
-
A two-tiered mechanism by which Cdc42 controls the localization and activation of an Arp2/3-activating motor complex in yeast.J Cell Biol. 2001 Oct 15;155(2):261-70. doi: 10.1083/jcb.200104094. Epub 2001 Oct 15. J Cell Biol. 2001. PMID: 11604421 Free PMC article.
-
Formins in development: orchestrating body plan origami.Biochim Biophys Acta. 2010 Feb;1803(2):207-25. doi: 10.1016/j.bbamcr.2008.09.016. Epub 2008 Oct 14. Biochim Biophys Acta. 2010. PMID: 18996154 Free PMC article. Review.
-
And the dead shall rise: actin and myosin return to the spindle.Dev Cell. 2011 Sep 13;21(3):410-9. doi: 10.1016/j.devcel.2011.07.018. Dev Cell. 2011. PMID: 21920311 Free PMC article. Review.
-
Gic2p may link activated Cdc42p to components involved in actin polarization, including Bni1p and Bud6p (Aip3p).Mol Cell Biol. 2000 Sep;20(17):6244-58. doi: 10.1128/MCB.20.17.6244-6258.2000. Mol Cell Biol. 2000. PMID: 10938101 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

