The boundaries of the silenced HMR domain in Saccharomyces cerevisiae
- PMID: 10090726
- PMCID: PMC316548
- DOI: 10.1101/gad.13.6.698
The boundaries of the silenced HMR domain in Saccharomyces cerevisiae
Abstract
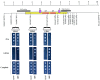
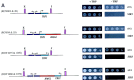
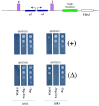
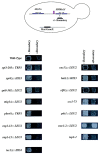
The chromosomes of eukaryotes are organized into structurally and functionally discrete domains that provide a mechanism to compact the DNA as well as delineate independent units of gene activity. It is believed that insulator/boundary elements separate these domains. Here we report the identification and characterization of boundary elements that flank the transcriptionally repressed HMR locus in the yeast Saccharomyces cerevisiae. Deletion of these boundary elements led to the spread of silenced chromatin, whereas the ectopic insertion of these elements between a silencer and a promoter blocked the repressive effects of the silencer on that promoter at HMR and at telomeres. Sequence analysis indicated that the boundary element contained a TY1 LTR, and a tRNA gene and mutational analysis has implicated the Smc proteins, which encode structural components of chromosomes, in boundary element function.
Figures





References
-
- Brand AH, Breeden L, Abraham J, Sternglanz R, Nasmyth K. Characterization of a ‘silencer’ in yeast: A DNA sequence with properties opposite to those of a transcriptional enhancer. Cell. 1985;41:41–48. - PubMed
-
- Chuang PT, Albertson DG, Meyer BJ. DPY-27: A chromosome condensation protein homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell. 1994;79:459–474. - PubMed
-
- Corces VG, Geyer PK. Interactions of retrotransposons with the host genome: The case of the gypsy element of Drosophila. Trends Genet. 1991;7:86–90. - PubMed
-
- Eisenmann DM, Arndt KM, Ricupero SL, Rooney JW, Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes & Dev. 1992;6:1319–1331. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
