Human pancreatic islets express mRNA species encoding two distinct catalytically active isoforms of group VI phospholipase A2 (iPLA2) that arise from an exon-skipping mechanism of alternative splicing of the transcript from the iPLA2 gene on chromosome 22q13.1
- PMID: 10092647
- PMCID: PMC3715997
- DOI: 10.1074/jbc.274.14.9607
Human pancreatic islets express mRNA species encoding two distinct catalytically active isoforms of group VI phospholipase A2 (iPLA2) that arise from an exon-skipping mechanism of alternative splicing of the transcript from the iPLA2 gene on chromosome 22q13.1
Abstract
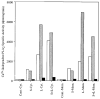
An 85-kDa Group VI phospholipase A2 enzyme (iPLA2) that does not require Ca2+ for catalysis has recently been cloned from three rodent species. A homologous 88-kDa enzyme has been cloned from human B-lymphocyte lines that contains a 54-amino acid insert not present in the rodent enzymes, but human cells have not previously been observed to express catalytically active iPLA2 isoforms other than the 88-kDa protein. We have cloned cDNA species that encode two distinct iPLA2 isoforms from human pancreatic islet RNA and a human insulinoma cDNA library. One isoform is an 85-kDa protein (short isoform of human iPLA2 (SH-iPLA2)) and the other an 88-kDa protein (long isoform of human iPLA2 (LH-iPLA2)). Transcripts encoding both isoforms are also observed in human promonocytic U937 cells. Recombinant SH-iPLA2 and LH-iPLA2 are both catalytically active in the absence of Ca2+ and inhibited by a bromoenol lactone suicide substrate, but LH-iPLA2 is activated by ATP, whereas SH-iPLA2 is not. The human iPLA2 gene has been found to reside on chromosome 22 in region q13.1 and to contain 16 exons represented in the LH-iPLA2 transcript. Exon 8 is not represented in the SH-iPLA2 transcript, indicating that it arises by an exon-skipping mechanism of alternative splicing. The amino acid sequence encoded by exon 8 of the human iPLA2 gene is proline-rich and shares a consensus motif of PX5PX8HHPX12NX4Q with the proline-rich middle linker domains of the Smad proteins DAF-3 and Smad4. Expression of mRNA species encoding two active iPLA2 isoforms with distinguishable catalytic properties in two different types of human cells demonstrated here may have regulatory or functional implications about the roles of products of the iPLA2 gene in cell biologic processes.
Figures

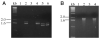


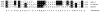



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

