Mutational analysis of the phoD promoter in Bacillus subtilis: implications for PhoP binding and promoter activation of Pho regulon promoters
- PMID: 10094677
- PMCID: PMC93612
- DOI: 10.1128/JB.181.7.2017-2025.1999
Mutational analysis of the phoD promoter in Bacillus subtilis: implications for PhoP binding and promoter activation of Pho regulon promoters
Abstract
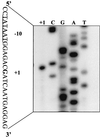
The PhoP-PhoR two-component regulatory system controls the phosphate deficiency response in B. subtilis. A number of Pho regulon genes which require PhoP approximately P for activation or repression have been identified. The studies reported here were initiated to understand the PhoP-DNA interaction necessary for Pho promoter regulation. The regulatory region of phoD was characterized in detail using oligo-directed mutagenesis, DNase I footprinting, and in vivo transcription assays. These data reveal basic principles of PhoP binding relevant to PhoP's interaction with other Pho regulon promoters. Our results show that: (i) a dimer of PhoP approximately P is able to bind two consensus repeats in a stable fashion; (ii) PhoP binding is highly cooperative within the core promoter region, which is located from -66 to -17 on the coding strand and contains four TT(A/T/C)ACA-like repeats; (iii) specific bases comprising the TT(A/T/C)ACA consensus are essential for transcriptional activation, but the specific base pairs of the intervening sequences separating the consensus repeats are not important for either PhoP binding or promoter activation; (iv) the spacing between two consensus repeats within a putative dimer binding site in the core region is important for both PhoP binding and promoter activation; (v) the exact spacing between two dimer binding sites within the core region is important for promoter activation but less so for PhoP binding affinity, as long as the repeats are on the same face of the helix; and (vi) the 5' secondary binding region is important for coordinated PhoP binding to the core binding region, making it nearly essential for promoter activation.
Figures






References
-
- Chesnut R S, Bookstein C, Hulett F M. Separate promoters direct expression of phoAIII, a member of the Bacillus subtilis alkaline phosphatase multigene family, during phosphate starvation and sporulation. Mol Microbiol. 1991;5:2181–2190. - PubMed
-
- Cutting S M, Vander Horn J A. Genetic analysis. In: Harwood C R, Cutting S M, editors. Molecular biological methods for bacillus. New York, N.Y: John Wiley; 1990. pp. 27–74.
-
- Eder S, Shi L, Jensen K, Yamane K, Hulett F M. A Bacillus subtilis secreted phosphodiesterase/alkaline phosphatase is the product of a Pho regulon gene, phoD. Microbiology. 1996;142:2041–2047. - PubMed
-
- Hanahan D. Techniques for transformation of E. coli. In: Glover D M, editor. DNA cloning II: a practical approach. Washington, D.C: IRL Press; 1985. pp. 109–135.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

