Role of bkdR, a transcriptional activator of the sigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis
- PMID: 10094682
- PMCID: PMC93617
- DOI: 10.1128/JB.181.7.2059-2066.1999
Role of bkdR, a transcriptional activator of the sigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis
Abstract
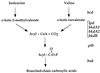
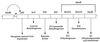
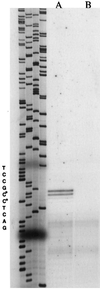
A new gene, bkdR (formerly called yqiR), encoding a regulator with a central (catalytic) domain was found in Bacillus subtilis. This gene controls the utilization of isoleucine and valine as sole nitrogen sources. Seven genes, previously called yqiS, yqiT, yqiU, yqiV, bfmBAA, bfmBAB, and bfmBB and now referred to as ptb, bcd, buk, lpd, bkdA1, bkdA2, and bkdB, are located downstream from the bkdR gene in B. subtilis. The products of these genes are similar to phosphate butyryl coenzyme A transferase, leucine dehydrogenase, butyrate kinase, and four components of the branched-chain keto acid dehydrogenase complex: E3 (dihydrolipoamide dehydrogenase), E1alpha (dehydrogenase), E1beta (decarboxylase), and E2 (dihydrolipoamide acyltransferase). Isoleucine and valine utilization was abolished in bcd and bkdR null mutants of B. subtilis. The seven genes appear to be organized as an operon, bkd, transcribed from a -12, -24 promoter. The expression of the bkd operon was induced by the presence of isoleucine or valine in the growth medium and depended upon the presence of the sigma factor SigL, a member of the sigma 54 family. Transcription of this operon was abolished in strains containing a null mutation in the regulatory gene bkdR. Deletion analysis showed that upstream activating sequences are involved in the expression of the bkd operon and are probably the target of bkdR. Transcription of the bkd operon is also negatively controlled by CodY, a global regulator of gene expression in response to nutritional conditions.
Figures


References
-
- Arantes O, Lereclus D. Construction of cloning vectors for Bacillus thuringiensis. Gene. 1991;108:115–119. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

