Transcriptional activation of ydeA, which encodes a member of the major facilitator superfamily, interferes with arabinose accumulation and induction of the Escherichia coli arabinose PBAD promoter
- PMID: 10094697
- PMCID: PMC93632
- DOI: 10.1128/JB.181.7.2185-2191.1999
Transcriptional activation of ydeA, which encodes a member of the major facilitator superfamily, interferes with arabinose accumulation and induction of the Escherichia coli arabinose PBAD promoter
Abstract
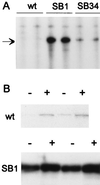
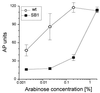
Induction of genes expressed from the arabinose PBAD promoter is very rapid and maximal at low arabinose concentrations. We describe here two mutations that interfere with the expression of genes cloned under arabinose control. Both mutations map to the ydeA promoter and stimulate ydeA transcription; overexpression of YdeA from a multicopy plasmid confers the same phenotype. One mutation is a large deletion that creates a more efficient -35 region (ATCACA changed to TTCACA), whereas the other affects the initiation site (TTTT changed to TGTT). The ydeA gene is expressed at extremely low levels in exponentially growing wild-type cells and is not induced by arabinose. Disruption of ydeA has no detectable effect on cell growth. Thus, ydeA appears to be nonessential under usual laboratory growth conditions. The ydeA gene encodes a membrane protein with 12 putative transmembrane segments. YdeA belongs to the largest family of bacterial secondary active transporters, the major facilitator superfamily, which includes antibiotic resistance exporters, Lac permease, and the nonessential AraJ protein. Intracellular accumulation of arabinose is strongly decreased in mutant strains overexpressing YdeA, suggesting that YdeA facilitates arabinose export. Consistent with this interpretation, very high arabinose concentrations can compensate for the negative effect of ydeA transcriptional activation. Our studies (i) indicate that YdeA, when transcriptionally activated, contributes to the control of the arabinose regulon and (ii) demonstrate a new way to modulate the kinetics of induction of cloned genes.
Figures



References
-
- Belin D. The use of RNA probes for the analysis of gene expression. Mol Biotechnol. 1997;7:153–163. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

