Design of highly specific cytotoxins by using trans-splicing ribozymes
- PMID: 10097066
- PMCID: PMC22323
- DOI: 10.1073/pnas.96.7.3507
Design of highly specific cytotoxins by using trans-splicing ribozymes
Abstract
We have designed ribozymes based on a self-splicing group I intron that can trans-splice exon sequences into a chosen RNA target to create a functional chimeric mRNA and provide a highly specific trigger for gene expression. We have targeted ribozymes against the coat protein mRNA of a widespread plant pathogen, cucumber mosaic virus. The ribozymes were designed to trans-splice the coding sequence of the diphtheria toxin A chain in frame with the viral initiation codon of the target sequence. Diphtheria toxin A chain catalyzes the ADP ribosylation of elongation factor 2 and can cause the cessation of protein translation. In a Saccharomyces cerevisiae model system, ribozyme expression was shown to specifically inhibit the growth of cells expressing the virus mRNA. A point mutation at the target splice site alleviated this ribozyme-mediated toxicity. Increasing the extent of base pairing between the ribozyme and target dramatically increased specific expression of the cytotoxin and reduced illegitimate toxicity in vivo. Trans-splicing ribozymes may provide a new class of agents for engineering virus resistance and therapeutic cytotoxins.
Figures
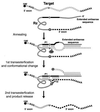
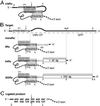



References
-
- Cech T R. Annu Rev Biochem. 1990;59:543–568. - PubMed
-
- Galloway-Salvo J L, Coetzee T, Belfort M. J Mol Biol. 1990;211:537–549. - PubMed
-
- Sullenger B A, Cech T R. Nature (London) 1994;371:619–622. - PubMed
-
- Jones J T, Lee S W, Sullenger B A. Nat Med. 1996;2:643–648. - PubMed
-
- Sarver N, Cairns S. Nat Med. 1996;2:641–642. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

