A cytosine analog that confers enhanced potency to antisense oligonucleotides
- PMID: 10097067
- PMCID: PMC22324
- DOI: 10.1073/pnas.96.7.3513
A cytosine analog that confers enhanced potency to antisense oligonucleotides
Abstract
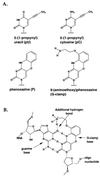
Antisense technology is based on the ability to design potent, sequence-specific inhibitors. The G-clamp heterocycle modification, a cytosine analog that clamps on to guanine by forming an additional hydrogen bond, was rationally designed to enhance oligonucleotide/RNA hybrid affinity. A single, context-dependent substitution of a G-clamp heterocycle into a 15-mer phosphorothioate oligodeoxynucleotide (S-ON) targeting the cyclin-dependent kinase inhibitor, p27(kip1), enhanced antisense activity as compared with a previously optimized C5-propynyl-modified p27(kip1) S-ON and functionally replaced 11 C5-propynyl modifications. Dose-dependent, sequence-specific antisense inhibition was observed at nanomolar concentrations of the G-clamp S-ONs. A single nucleotide mismatch between the G-clamp S-ON and the p27(kip1) mRNA reduced the potency of the antisense ON by five-fold. A 2-base-mismatch S-ON eliminated antisense activity, confirming the sequence specificity of G-clamp-modified S-ONs. The G-clamp-substituted p27(kip1) S-ON activated RNase H-mediated cleavage and demonstrated increased in vitro binding affinity for its RNA target compared with conventional 15-mer S-ONs. Furthermore, incorporation of a single G-clamp modification into a previously optimized 20-mer phosphorothioate antisense S-ON targeting c-raf increased the potency of the S-ON 25-fold. The G-clamp heterocycle is a potent, mismatch-sensitive, automated synthesizer-compatible antisense S-ON modification that will have important applications in the elucidation of gene function, the validation of gene targets, and the development of more potent antisense-based pharmaceuticals.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

