Crystal structure of human type II inosine monophosphate dehydrogenase: implications for ligand binding and drug design
- PMID: 10097070
- PMCID: PMC22327
- DOI: 10.1073/pnas.96.7.3531
Crystal structure of human type II inosine monophosphate dehydrogenase: implications for ligand binding and drug design
Abstract
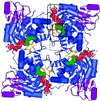
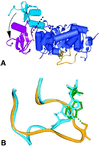
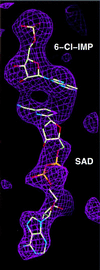
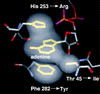
Inosine monophosphate dehydrogenase (IMPDH) controls a key metabolic step in the regulation of cell growth and differentiation. This step is the NAD-dependent oxidation of inosine 5' monophosphate (IMP) to xanthosine 5' monophosphate, the rate-limiting step in the synthesis of the guanine nucleotides. Two isoforms of IMPDH have been identified, one of which (type II) is significantly up- regulated in neoplastic and differentiating cells. As such, it has been identified as a major target in antitumor and immunosuppressive drug design. We present here the 2.9-A structure of a ternary complex of the human type II isoform of IMPDH. The complex contains the substrate analogue 6-chloropurine riboside 5'-monophosphate (6-Cl-IMP) and the NAD analogue selenazole-4-carboxamide adenine dinucleotide, the selenium derivative of the active metabolite of the antitumor drug tiazofurin. The enzyme forms a homotetramer, with the dinucleotide binding at the monomer-monomer interface. The 6 chloro-substituted purine base is dehalogenated, forming a covalent adduct at C6 with Cys-331. The dinucleotide selenazole base is stacked against the 6-Cl-IMP purine ring in an orientation consistent with the B-side stereochemistry of hydride transfer seen with NAD. The adenosine end of the ligand interacts with residues not conserved between the type I and type II isoforms, suggesting strategies for the design of isoform-specific agents.
Figures
Similar articles
-
Crystal structure of a ternary complex of Tritrichomonas foetus inosine 5'-monophosphate dehydrogenase: NAD+ orients the active site loop for catalysis.Biochemistry. 2002 Nov 5;41(44):13309-17. doi: 10.1021/bi0203785. Biochemistry. 2002. PMID: 12403633
-
Crystal structure at 2.4 A resolution of Borrelia burgdorferi inosine 5'-monophosphate dehydrogenase: evidence of a substrate-induced hinged-lid motion by loop 6.Biochemistry. 2000 Apr 18;39(15):4533-42. doi: 10.1021/bi992645l. Biochemistry. 2000. PMID: 10758003
-
IMP dehydrogenase : structural aspects of inhibitor binding.Curr Med Chem. 1999 Jul;6(7):519-36. Curr Med Chem. 1999. PMID: 10390598 Review.
-
Probing the active site of human IMP dehydrogenase using halogenated purine riboside 5'-monophosphates and covalent modification reagents.Biochemistry. 1994 Feb 22;33(7):1760-5. doi: 10.1021/bi00173a019. Biochemistry. 1994. PMID: 7906543
-
The structure of inosine 5'-monophosphate dehydrogenase and the design of novel inhibitors.Immunopharmacology. 2000 May;47(2-3):163-84. doi: 10.1016/s0162-3109(00)00193-4. Immunopharmacology. 2000. PMID: 10878288 Review.
Cited by
-
Polymorphisms in type I and II inosine monophosphate dehydrogenase genes and association with clinical outcome in patients on mycophenolate mofetil.Pharmacogenet Genomics. 2010 Sep;20(9):537-43. doi: 10.1097/FPC.0b013e32833d8cf5. Pharmacogenet Genomics. 2010. PMID: 20679962 Free PMC article.
-
Identification of the human mitochondrial linoleoyl-coenzyme A monolysocardiolipin acyltransferase (MLCL AT-1).J Biol Chem. 2009 Oct 30;284(44):30360-71. doi: 10.1074/jbc.M109.048322. Epub 2009 Sep 8. J Biol Chem. 2009. PMID: 19737925 Free PMC article.
-
The cystathionine-β-synthase domains on the guanosine 5''-monophosphate reductase and inosine 5'-monophosphate dehydrogenase enzymes from Leishmania regulate enzymatic activity in response to guanylate and adenylate nucleotide levels.Mol Microbiol. 2016 Jun;100(5):824-40. doi: 10.1111/mmi.13352. Epub 2016 Mar 10. Mol Microbiol. 2016. PMID: 26853689 Free PMC article.
-
IMPDH2's Central Role in Cellular Growth and Diseases: A Potential Therapeutic Target.Cell Prolif. 2025 Jun;58(6):e70031. doi: 10.1111/cpr.70031. Epub 2025 Apr 19. Cell Prolif. 2025. PMID: 40251939 Free PMC article. Review.
-
Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.Biochemistry. 2012 Aug 7;51(31):6148-63. doi: 10.1021/bi300511w. Epub 2012 Jul 25. Biochemistry. 2012. PMID: 22788966 Free PMC article.
References
-
- Weber G, Prajda N, Abonyi M, Look K Y, Tricot G. Anticancer Res. 1996;16:3313–3322. - PubMed
-
- Wu J C. Perspect Drug Discovery Des. 1994;2:185–204.
-
- Franklin T J, Edwards G, Hedge P. Adv Exp Med Biol. 1994;370:155–160. - PubMed
-
- Weber G, Prajda N, Jackson R C. Adv Enzyme Regul. 1976;14:3–24. - PubMed
-
- Jackson R C, Weber G, Morris H P. Nature (London) 1975;256:331–333. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

